4BDB
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4LQ3
 
 | | Crystal structure of human norovirus RNA-dependent RNA-polymerase bound to the inhibitor PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, 5'-R(P*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Milani, M, Tarantino, D, Mastrangelo, E, Croci, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Antiviral Res., 102, 2014
|
|
3RV8
 
 | | Structure of a M. tuberculosis Salicylate Synthase, MbtI, in Complex with an Inhibitor with Cyclopropyl R-Group | | Descriptor: | 3-{[(E)-1-carboxy-2-cyclopropylethenyl]oxy}-2-hydroxybenzoic acid, 3-{[(Z)-1-carboxy-2-cyclopropylethenyl]oxy}-2-hydroxybenzoic acid, Isochorismate synthase/isochorismate-pyruvate lyase mbtI | | Authors: | Chi, G, Bulloch, E.M.M, Manos-Turvey, A, Payne, R.J, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis
Biochemistry, 51, 2012
|
|
7DYQ
 
 | | Crystal structure of histone lysine demethylase 4D (KDM4D) in complex with the inhibitor 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile | | Descriptor: | 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile, FE (III) ION, Lysine-specific demethylase 4D | | Authors: | Wang, T, Yang, L. | | Deposit date: | 2021-01-22 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of histone lysine demethylase 4D (KDM4D) in complex with the inhibitor 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile
To Be Published
|
|
3RV6
 
 | | Structure of a M. tuberculosis Salicylate Synthase, MbtI, in Complex with an Inhibitor with Phenyl R-Group | | Descriptor: | 3-{[(E)-1-carboxy-2-phenylethenyl]oxy}-2-hydroxybenzoic acid, 3-{[(Z)-1-carboxy-2-phenylethenyl]oxy}-2-hydroxybenzoic acid, Isochorismate synthase/isochorismate-pyruvate lyase mbtI, ... | | Authors: | Chi, G, Bulloch, E.M.M, Manos-Turvey, A, Payne, R.J, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis
Biochemistry, 51, 2012
|
|
3DHW
 
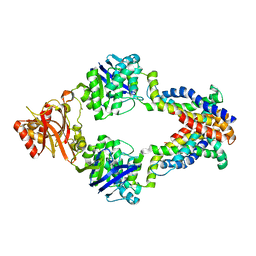 | | Crystal structure of methionine importer MetNI | | Descriptor: | D-methionine transport system permease protein metI, Methionine import ATP-binding protein metN | | Authors: | Rees, D.C, Kaiser, J.T, Kadaba, N.S, Johnson, E, Lee, A.T. | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation.
Science, 321, 2008
|
|
3CZ0
 
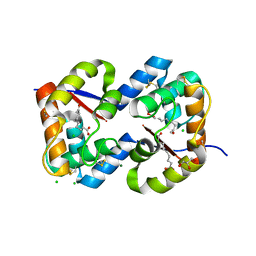 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3MDW
 
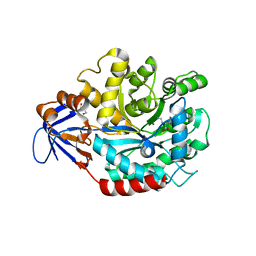 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | GLYCEROL, N-[(E)-iminomethyl]-L-aspartic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8979 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
4KFB
 
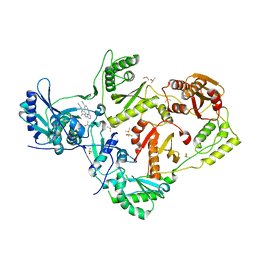 | | HIV-1 reverse transcriptase with bound fragment at NNRTI adjacent site | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, P51 RT, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-Ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
1YWL
 
 | |
4DVG
 
 | |
2AWQ
 
 | |
3TH8
 
 | | Structure of E. coli undecaprenyl diphosphate synthase complexed with BPH-1063 | | Descriptor: | (2Z)-4-({3-[3-(hexyloxy)phenyl]propyl}amino)-2-hydroxy-4-oxobut-2-enoic acid, Undecaprenyl pyrophosphate synthase | | Authors: | Cao, R, Zhu, W, Zhang, Y, Oldfield, E. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
3PAA
 
 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 8.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
4BWL
 
 | | Structure of the Y137A mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate, N-acetyl-D-mannosamine and N- acetylneuraminic acid | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, N-ACETYLNEURAMINATE LYASE, ... | | Authors: | Campeotto, I, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2013-07-03 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Reaction Mechanism of N-Acetylneuraminic Acid Lyase Revealed by a Combination of Crystallography, Qm/Mm Simulation and Mutagenesis.
Acs Chem.Biol., 9, 2014
|
|
2FEM
 
 | | Mutant R188M of the Cytidine Monophosphate Kinase From E. Coli | | Descriptor: | Cytidylate kinase | | Authors: | Ofiteru, A, Bucurenci, N, Alexov, E, Bertrand, T, Briozzo, P, Munier-Lehmann, H, Tourneux, L, Barzu, O, Gilles, A.M. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional consequences of single amino acid substitutions in the pyrimidine base binding pocket of Escherichia coli CMP kinase.
Febs J., 274, 2007
|
|
1G7O
 
 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | Descriptor: | GLUTAREDOXIN 2 | | Authors: | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-11-10 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
4O4R
 
 | | Murine Norovirus RdRp in complex with PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, RNA-dependent-RNA-polymerase, SULFATE ION | | Authors: | Croci, R, Tarantino, D, Milani, M, Pezzullo, M, Bolognesi, M, Mastrangelo, E. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PPNDS inhibits murine Norovirus RNA-dependent RNA-polymerase mimicking two RNA stacking bases.
Febs Lett., 588, 2014
|
|
3QJH
 
 | | The crystal structure of the 5c.c7 TCR | | Descriptor: | 5c.c7 alpha chain, 5c.c7 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QJF
 
 | | Crystal structure of the 2B4 TCR | | Descriptor: | 2B4 alpha chain, 2B4 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
2FEO
 
 | | Mutant R188M of The Cytidine Monophosphate Kinase from E. coli complexed with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Cytidylate kinase, SULFATE ION | | Authors: | Ofiteru, A, Bucurenci, N, Alexov, E, Bertrand, T, Briozzo, P, Munier-Lehmann, H, Tourneux, L, Barzu, O, Gilles, A.M. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional consequences of single amino acid substitutions in the pyrimidine base binding pocket of Escherichia coli CMP kinase.
Febs J., 274, 2007
|
|
3LXG
 
 | | Crystal structure of rat phosphodiesterase 10A in complex with ligand WEB-3 | | Descriptor: | 2-methoxy-6,7-dimethyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Mosbacher, T, Jestel, A, Steinbacher, S. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,5-a]pyrido[3,2-e]pyrazines as a new class of phosphodiesterase 10A inhibitiors.
J.Med.Chem., 53, 2010
|
|
4BEN
 
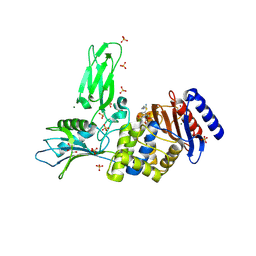 | | R39-imipenem Acyl-enzyme crystal structure | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, ... | | Authors: | Van Elder, D, Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-03-11 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of R39-Imipenem Acyl-Enzyme.
To be Published
|
|
3MEG
 
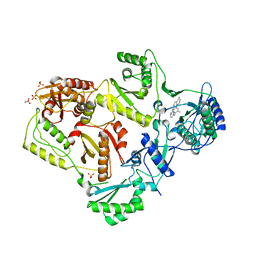 | | HIV-1 K103N Reverse Transcriptase in Complex with TMC278 | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
4QMT
 
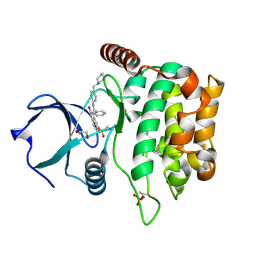 | | MST3 in complex with HESPERADIN | | Descriptor: | 1,2-ETHANEDIOL, N-[2-OXO-3-((E)-PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]IMINO}METHYL)-2,6-DIHYDRO-1H-INDOL-5-YL]ETHANESULFONAMIDE, Serine/threonine-protein kinase 24 | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
