5T8V
 
 | | Chaetomium thermophilum cohesin loader SCC2, C-terminal fragment | | Descriptor: | CITRIC ACID, Putative uncharacterized protein | | Authors: | Tomchick, D.R, Yu, H, Kikuchi, S, Ouyang, Z, Borek, D, Otwinowski, Z. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2C7M
 
 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | Descriptor: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | Authors: | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | Deposit date: | 2005-11-25 | | Release date: | 2006-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin.
Cell, 124, 2006
|
|
5V3N
 
 | | Structure of S. cerevisiae Ulp2-Tof2-Csm1 complex | | Descriptor: | Monopolin complex subunit CSM1, Ulp2p,Topoisomerase 1-associated factor 2 chimera | | Authors: | SIngh, N, Corbett, K.D. | | Deposit date: | 2017-03-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity.
Genes Dev., 31, 2017
|
|
2DYD
 
 | |
5JYQ
 
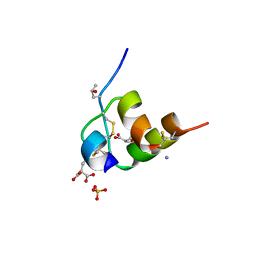 | | Structure of Conus Geographus insulin G1 | | Descriptor: | Insulin 1, Insulin 1b, SULFATE ION | | Authors: | Lawrence, M.C, Menting, J, Norton, R.S, Safavi-Hemami, H. | | Deposit date: | 2016-05-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5VAX
 
 | |
5UZ9
 
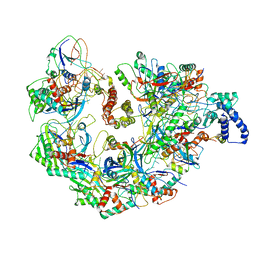 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
4BJS
 
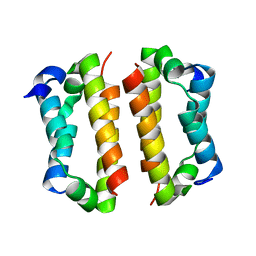 | | Crystal structure of the Rif1 C-terminal domain (Rif1-CTD) from Saccharomyces cerevisiae | | Descriptor: | TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Bunker, R.D, Shi, T, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
5VAY
 
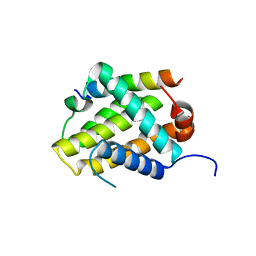 | |
5UNA
 
 | | Fragment of 7SK snRNA methylphosphate capping enzyme | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, unidentified peptide section/fragment | | Authors: | Wu, H, Tempel, W, Dombrovski, L, McCarthy, A.A, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-30 | | Release date: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment of 7SK snRNA methylphosphate capping enzyme
To Be Published
|
|
5TM9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, (E)-3-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)acrylic acid | | Descriptor: | 3-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}prop-2-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMR
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Cyclofenil-ASC derivative, ethyl (E)-3-(4-(cyclohexylidene(4-hydroxyphenyl)methyl)phenyl)acrylate | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, ethyl 3-{4-[cyclohexylidene(4-hydroxyphenyl)methyl]phenyl}prop-2-enoate | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S,E)-17-((4-isopropylphenyl)imino)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol | | Descriptor: | (9beta,13alpha,17Z)-17-{[4-(propan-2-yl)phenyl]imino}estra-1,3,5(10)-trien-3-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM1
 
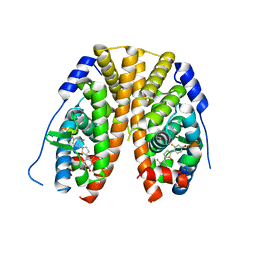 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,5-bis(2-fluoro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | 2,5-bis(2-fluoro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5U4M
 
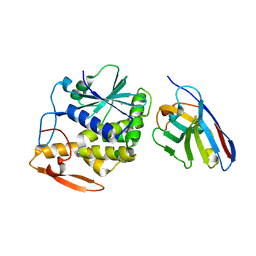 | | RTA-V1C7-G29R-no_salt | | Descriptor: | CHLORIDE ION, Ricin, V1C7 VHH antibody | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Using homology modeling to interrogate binding affinity in neutralization of ricin toxin by a family of single domain antibodies.
Proteins, 85, 2017
|
|
5U34
 
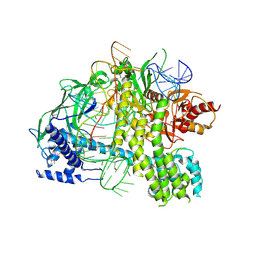 | | Crystal structure of AacC2c1-sgRNA binary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, sgRNA | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.255 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5TQU
 
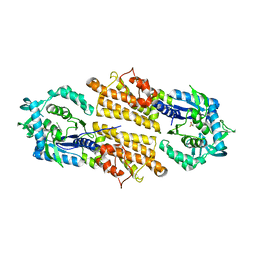 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor | | Descriptor: | 2-[2-[[3-(1~{H}-benzimidazol-2-ylamino)propylamino]methyl]-4,6-bis(chloranyl)indol-1-yl]ethanol, GLYCEROL, METHIONINE, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Cells to Mice to Target: Characterization of NEU-1053 (SB-443342) and Its Analogues for Treatment of Human African Trypanosomiasis.
ACS Infect Dis, 3, 2017
|
|
5U33
 
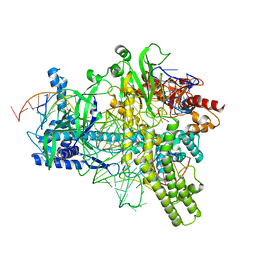 | | Crystal structure of AacC2c1-sgRNA-extended non-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5UE5
 
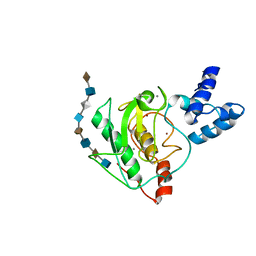 | | proMMP-7 with heparin octasaccharide bound to the catalytic domain | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
5UE2
 
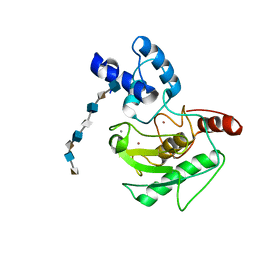 | | proMMP-7 with heparin octasaccharide bridging between domains | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
5V1A
 
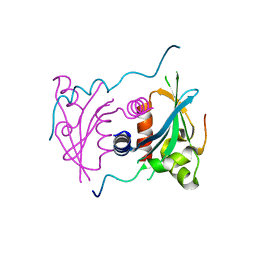 | | Structure of S. cerevisiae Ulp2:Csm1 complex | | Descriptor: | Monopolin complex subunit CSM1, Ubiquitin-like-specific protease 2 | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity.
Genes Dev., 31, 2017
|
|
4EQL
 
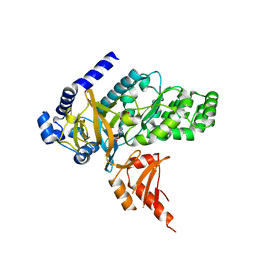 | | Crystal Structure of GH3.12 in complex with AMP and salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Westfall, C, Zubieta, C, Nanao, M, Herrmann, J, Jez, J. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
5TLL
 
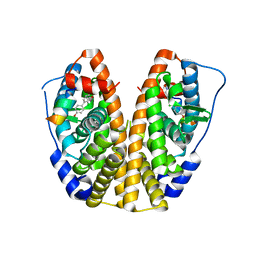 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with (E)-2-chloro-4'-hydroxy-4-((hydroxyiminio)methyl)-[1,1'-biphenyl]-3-olate | | Descriptor: | 2-chloro-4-[(E)-(hydroxyimino)methyl][1,1'-biphenyl]-3,4'-diol, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TLT
 
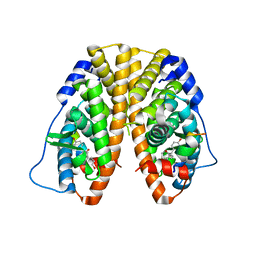 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with octane-1,8-diyl bis(2,3-bis(4-hydroxyphenyl)pentanoate) | | Descriptor: | 8-{[2,3-bis(4-hydroxyphenyl)pentanoyl]oxy}octyl (2R,3S)-2,3-bis(4-hydroxyphenyl)pentanoate, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5T9T
 
 | | Protocadherin Gamma B2 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin gamma B2-alpha C, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
