3P88
 
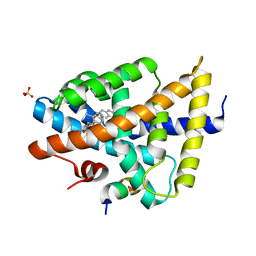 | | FXR bound to isoquinolinecarboxylic acid | | Descriptor: | 7-(4-{[3-(2,6-dimethylphenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)isoquinoline-3-carboxylic acid, Farnesoid X receptor, Nuclear receptor coactivator 1, ... | | Authors: | Madauss, K.P, Williams, S.P, Deaton, D.N. | | Deposit date: | 2010-10-13 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Heteroaryl replacements of the naphthalene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4L9E
 
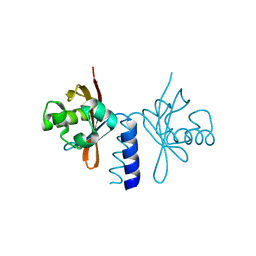 | | Structure of PpsR Q-PAS1 from Rb. sphaeroides | | Descriptor: | Transcriptional regulator, PpsR | | Authors: | Heintz, U, Meinhart, A, Schlichting, I, Winkler, A. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multi-PAS domain-mediated protein oligomerization of PpsR from Rhodobacter sphaeroides.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3EYZ
 
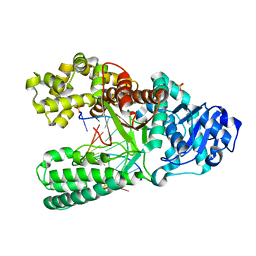 | | Cocrystal structure of Bacillus fragment DNA polymerase I with duplex DNA (open form) | | Descriptor: | 5'-D(*DAP*DTP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DAP*DGP*DGP*DA)-3', 5'-D(*DCP*DCP*DTP*DGP*DAP*DCP*DTP*DCP*DGP*DC)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Wu, E.Y, Golosov, A.A, Karplus, M, Beese, L.S. | | Deposit date: | 2008-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of the Translocation Step in DNA Replication by DNA Polymerase I: A Computer Simulation Analysis.
Structure, 18, 2010
|
|
4IUS
 
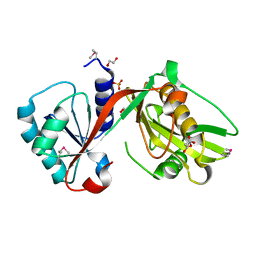 | | GCN5-related N-acetyltransferase from Kribbella flavida. | | Descriptor: | 1,2-ETHANEDIOL, GCN5-related N-acetyltransferase, MALONATE ION, ... | | Authors: | Osipiuk, J, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-21 | | Release date: | 2013-01-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | GCN5-related N-acetyltransferase from Kribbella flavida.
To be Published
|
|
3FOM
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide IQQSIERL | | Descriptor: | 8 residue synthetic peptide, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Brims, D.R, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Malashkevich, V.N, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
2ICY
 
 | | Crystal Structure of a Putative UDP-glucose Pyrophosphorylase from Arabidopsis Thaliana with Bound UDP-glucose | | Descriptor: | DIMETHYL SULFOXIDE, Probable UTP-glucose-1-phosphate uridylyltransferase 2, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
3FPE
 
 | | Crystal Structure of MtNAS in complex with thermonicotianamine | | Descriptor: | BROMIDE ION, N-[(3S)-3-{[(3S)-3-amino-3-carboxypropyl]amino}-3-carboxypropyl]-L-glutamic acid, Putative uncharacterized protein | | Authors: | Dreyfus, C, Pignol, D, Arnoux, P. | | Deposit date: | 2009-01-05 | | Release date: | 2009-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of iterative substrate translocations during nicotianamine synthesis in Archaea
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F0V
 
 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-(2,6-dimethylphenyl)phenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3S)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-26 | | Release date: | 2009-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic Complexes of Wildtype and Mutant MRSA DHFR Reveal Interactions for Lead Design
To be Published
|
|
4L4O
 
 | | The crystal structure of CbXyn10B in native form | | Descriptor: | Endo-1,4-beta-xylanase, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | An, J, Feng, Y, Wu, G. | | Deposit date: | 2013-06-08 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of CbXyn10B from Caldicellulosiruptor bescii and its mutant(E139A) in complex with xylotriose
To be Published
|
|
4KWG
 
 | | Crystal Structure Analysis of ALDH2+ALDiB13 | | Descriptor: | 1,2-ETHANEDIOL, 7-bromo-5-methyl-1H-indole-2,3-dione, Aldehyde dehydrogenase, ... | | Authors: | Hurley, T.D, Kimble-Hill, A.C. | | Deposit date: | 2013-05-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole-2,3-diones.
J.Med.Chem., 57, 2014
|
|
3FPL
 
 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Felix, F, Goihberg, E, Shimon, L, Burstein, Y. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
4ISN
 
 | | Crystal Structure of Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | GLUTATHIONE, Kunitz-type protease inhibitor 1, Suppressor of tumorigenicity 14 protein, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
3F13
 
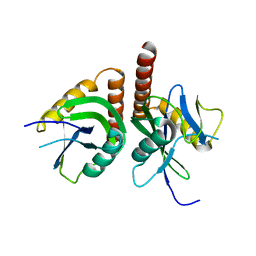 | | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum | | Descriptor: | putative nudix hydrolase family member | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum
To be Published
|
|
3FPZ
 
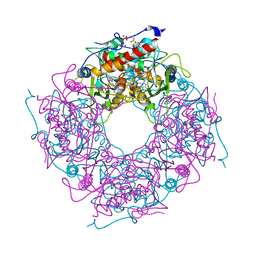 | | Saccharomyces cerevisiae THI4p is a suicide thiamin thiazole synthase | | Descriptor: | ADENOSINE DIPHOSPHATE 5-(BETA-ETHYL)-4-METHYL-THIAZOLE-2-CARBOXYLIC ACID, SULFATE ION, Thiazole biosynthetic enzyme | | Authors: | Bale, S, Chatterjee, A, Dorrestein, P.C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase.
Nature, 478, 2011
|
|
4IZX
 
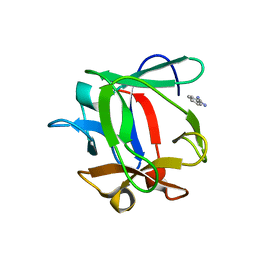 | | Macrolepiota procera ricin B-like lectin (MPL) in complex with lactose | | Descriptor: | BENZAMIDINE, Ricin B-like lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Renko, M, Zurga, S, Sabotic, J, Pohleven, J, Kos, J, Turk, D. | | Deposit date: | 2013-01-30 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Macrolepiota procera ricin B-like lectin (MPL) in complex with lactose
To be Published
|
|
3EXH
 
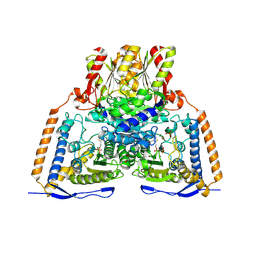 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
4KZ4
 
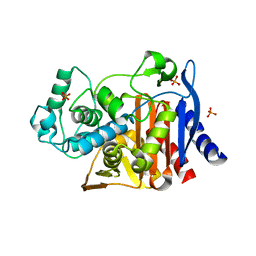 | | Crystal structure of AmpC beta-lactamase in complex with fragment 60 (2-[(propylsulfonyl)amino]benzoic acid) | | Descriptor: | 2-[(propylsulfonyl)amino]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
7TLA
 
 | |
4IV8
 
 | |
3OJY
 
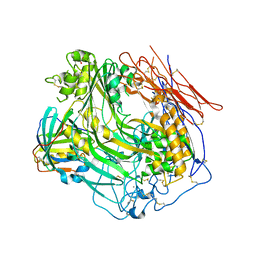 | | Crystal Structure of Human Complement Component C8 | | Descriptor: | CALCIUM ION, Complement component C8 alpha chain, Complement component C8 beta chain, ... | | Authors: | Lovelace, L.L, Cooper, C.L, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2010-08-23 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement.
J.Biol.Chem., 286, 2011
|
|
3F0M
 
 | | Crystal structure of radical D105N Synechocystis sp. PcyA | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Kohler, A.C, Gae, D.D. | | Deposit date: | 2008-10-25 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
3F0U
 
 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-phenylphenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3R)-3-(5-methoxybiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-26 | | Release date: | 2009-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.
J.Mol.Biol., 387, 2009
|
|
4L6X
 
 | |
4IX1
 
 | | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205 | | Descriptor: | PHOSPHATE ION, hypothetical protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205
To be Published
|
|
3P5N
 
 | |
