7ZW6
 
 | | Oligomeric structure of SynDLP | | Descriptor: | Slr0869 protein | | Authors: | Gewehr, L, Junglas, B, Jilly, R, Franz, J, Wenyu, E.Z, Weidner, T, Bonn, M, Sachse, C, Schneider, D. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | SynDLP is a dynamin-like protein of Synechocystis sp. PCC 6803 with eukaryotic features.
Nat Commun, 14, 2023
|
|
7ZW4
 
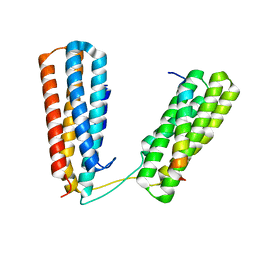 | | Crystal structure of Talin R7R8 domains with Caskin-2 LD-peptide | | Descriptor: | Caskin-2, Talin-1 | | Authors: | Celie, P.H.N, Joosten, R.P, Sonnenberg, A, Perrakis, A. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Caskin2 is a novel talin- and Abi1-binding protein that promotes cell motility.
J.Cell.Sci., 137, 2024
|
|
7ZW3
 
 | |
7ZW2
 
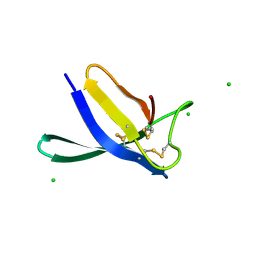 | | Penicillium expansum antifungal protein B | | Descriptor: | Antifungal protein, CHLORIDE ION | | Authors: | Gallego del Sol, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
7ZW1
 
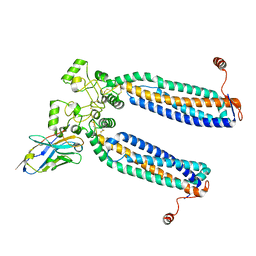 | | Human PRPH2-ROM1 hetero-dimer | | Descriptor: | Nanobody, Peripherin-2, Rod outer segment membrane protein 1 | | Authors: | El Mazouni, D, Gros, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of peripherin-2 and ROM1 suggest multiple roles in photoreceptor membrane morphogenesis.
Sci Adv, 8, 2022
|
|
7ZVZ
 
 | |
7ZVU
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZVR
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) complexed with zeaxanthin | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZVQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, S206V mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZVP
 
 | |
7ZVN
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 1.87 A resolution | | Descriptor: | 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, CALCIUM ION, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
7ZVL
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-16 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZVH
 
 | | Penicillium expansum antifungal protein B | | Descriptor: | Antifungal protein, CHLORIDE ION | | Authors: | Gallego del Sol, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
7ZVF
 
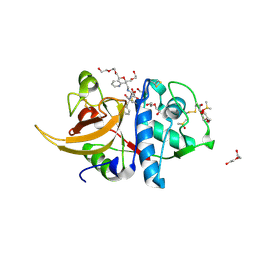 | | Crystal structure of human cathepsin L in complex with covalently bound CLIK148 | | Descriptor: | (2S)-N-[(2S)-1-(dimethylamino)-1-oxidanylidene-3-phenyl-propan-2-yl]-2-oxidanyl-N'-(2-pyridin-2-ylethyl)butanediamide, 1,2-ETHANEDIOL, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Tsuge, H, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
7ZVE
 
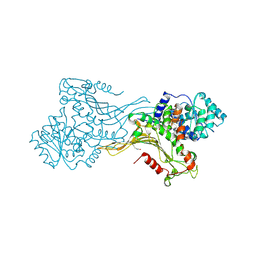 | | K403 acetylated glucose-6-phosphate dehydrogenase (G6PD) | | Descriptor: | COPPER (II) ION, GLYCEROL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wu, F, Muskat, N.H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
7ZVD
 
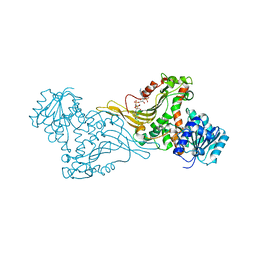 | | K89 acetylated glucose-6-phosphate dehydrogenase (G6PD) in a complex with structural NADP+ | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wu, F, Muskat, N.H, Nudelman, H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
7ZVC
 
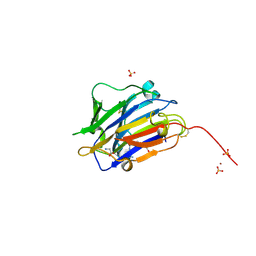 | | Second crystal form of the mature glutamic-class prolyl-endopeptidase neprosin at 1.85 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptidase, GLY-GLY-GLY-GLY, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Del Amo-Maestro, L, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZVB
 
 | | Crystal Structure of the mature form of the glutamic-class prolyl-endopeptidase neprosin at 2.35 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptidase, TRIETHYLENE GLYCOL, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZVA
 
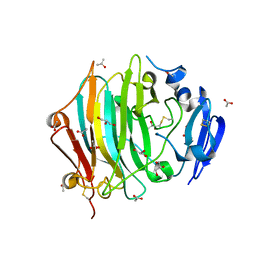 | | Crystal Structure of the native zymogen form of the glutamic-class prolyl-endopeptidase neprosin at 1.80 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C-terminal peptidase, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZV9
 
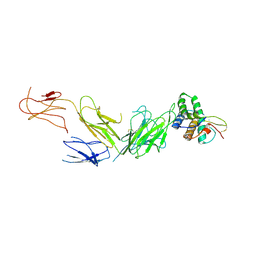 | |
7ZV8
 
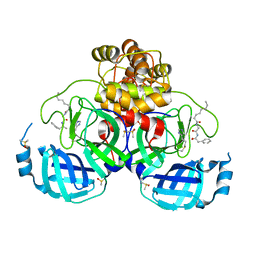 | | Crystal structure of SARS Cov-2 main protease in complex with an inhibitor 58 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Rahimova, R, Di Micco, S, Marquez, J.A. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Rational design of the zonulin inhibitor AT1001 derivatives as potential anti SARS-CoV-2.
Eur.J.Med.Chem., 244, 2022
|
|
7ZV7
 
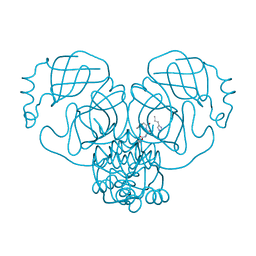 | |
7ZV5
 
 | |
7ZV3
 
 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0472 | | Descriptor: | 4-(3-chloro-2-phenethylphenyl)-1H-1,2,3-triazole, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2022-05-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structure-based optimization of type III indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7ZV2
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
