2PK4
 
 | |
2PUQ
 
 | |
2AER
 
 | | Crystal Structure of Benzamidine-Factor VIIa/Soluble Tissue Factor complex. | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bajaj, S.P, Schmidt, A.E, Padmanabhan, K, Bajaj, M.S, Liesum, A, Dumas, J, Prevost, D, Schreuder, H. | | Deposit date: | 2005-07-23 | | Release date: | 2006-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | High Resolution Structures of p-Aminobenzamidine- and Benzamidine-VIIa/Soluble Tissue Factor: Unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+ and Zn2+ sites in factor VIIa
J.Biol.Chem., 281, 2006
|
|
2A2Q
 
 | | Complex of Active-site Inhibited Human Coagulation Factor VIIa with Human Soluble Tissue Factor in the Presence of Ca2+, Mg2+, Na+, and Zn2+ | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII, ... | | Authors: | Bajaj, S.P, Bajaj, M, Schmidt, A.E, Padmanabhan, K. | | Deposit date: | 2005-06-22 | | Release date: | 2006-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of p-aminobenzamidine- and benzamidine-VIIa/soluble tissue factor: unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+, and Zn2+ sites in factor VIIa.
J.Biol.Chem., 281, 2006
|
|
1CEA
 
 | |
1CEB
 
 | |
4V08
 
 | | Inhibited dimeric pseudorabies virus protease pUL26N at 2 A resolution | | Descriptor: | CHLORIDE ION, DIISOPROPYL PHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-09-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
6ROJ
 
 | | Cryo-EM structure of the activated Drs2p-Cdc50p | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
7PEM
 
 | | Cryo-EM structure of phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Timcenko, M, Wang, Y, Lyons, J.A, Nissen, P, Lindorff-Larsen, K. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate Transport and Specificity in a Phospholipid Flippase
To Be Published
|
|
7OH4
 
 | | Cryo-EM structure of Drs2p-Cdc50p in the E1 state with PI4P and Mg2+ bound | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Dieudonne, T, Montigny, C, Boesen, T, Lyons, J.A, Lenoir, G, Nissen, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase
J.Mol.Biol., 2021
|
|
7OH7
 
 | | Cryo-EM structure of Drs2p-Cdc50p in the E1-AMPPCP state with PI4P bound | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | Authors: | Timcenko, M, Dieudonne, T, Montigny, C, Boesen, T, Lyons, J.A, Lenoir, G, Nissen, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase
J.Mol.Biol., 2021
|
|
7OH6
 
 | | Cryo-EM structure of Drs2p-Cdc50p in the [PS]E2-AlFx state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Dieudonne, T, Montigny, C, Boesen, T, Lyons, J.A, Lenoir, G, Nissen, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase
J.Mol.Biol., 2021
|
|
7OH5
 
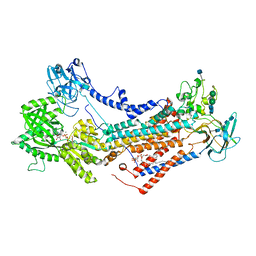 | | Cryo-EM structure of Drs2p-Cdc50p in the E1-AlFx-ADP state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Dieudonne, T, Montigny, C, Boesen, T, Lyons, J.A, Lenoir, G, Nissen, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase
J.Mol.Biol., 2021
|
|
5NJ6
 
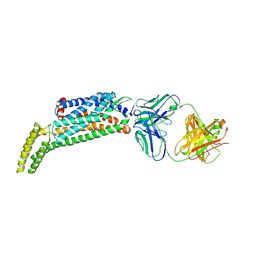 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | Descriptor: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
4JN1
 
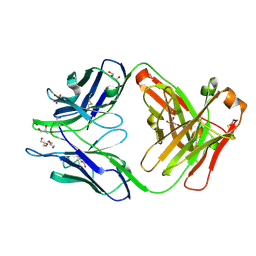 | | An Antidote for Dabigatran | | Descriptor: | GLYCEROL, anti-dabigatran Fab1, heavy chain, ... | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2013-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A specific antidote for dabigatran: functional and structural characterization.
Blood, 121, 2013
|
|
4JN2
 
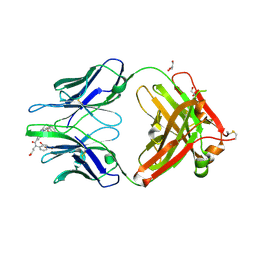 | | An Antidote for Dabigatran | | Descriptor: | GLYCEROL, N-[(2-{[(4-carbamimidoylphenyl)amino]methyl}-1-methyl-1H-benzimidazol-5-yl)carbonyl]-N-pyridin-2-yl-beta-alanine, anti dabigatran Fab | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2013-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A specific antidote for dabigatran: functional and structural characterization.
Blood, 121, 2013
|
|
8G6I
 
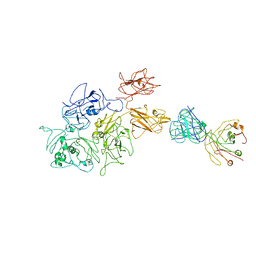 | | Coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor VIII chimera from human and pig, NB33 heavy chain, ... | | Authors: | Childers, K.C, Davulcu, O, Haynes, R.M, Lollar, P, Doering, C.B, Coxon, C.H, Spiegel, P.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor.
Blood, 142, 2023
|
|
8A8G
 
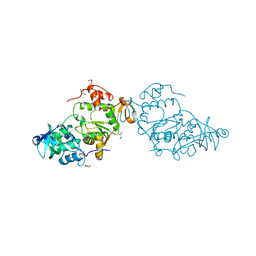 | |
8A8H
 
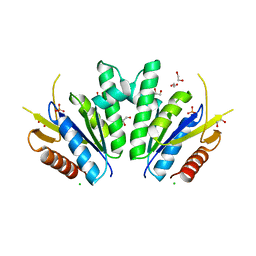 | |
8A8D
 
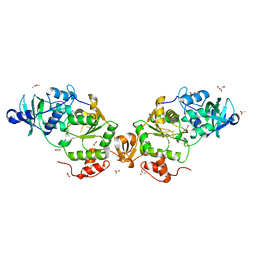 | |
7OQ4
 
 | | Cryo-EM structure of the ATV RNAP Inhibitory Protein (RIP) bound to the DNA-binding channel of the host's RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-02 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
5FS6
 
 | |
6MV4
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR IXa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schreuder, H.A, Liesum, A, Bajaj, S.P. | | Deposit date: | 2018-10-24 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Sodium-site in serine protease domain of human coagulation factor IXa: evidence from the crystal structure and molecular dynamics simulations study.
J. Thromb. Haemost., 17, 2019
|
|
6QAF
 
 | | Crystal structure of human Arginase-1 at pH 9.0 in complex with CB-1158/INCB001158 | | Descriptor: | Arginase-1, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Grobben, Y, Uitdehaag, J.C.M, Tabak, W.W.A, Zaman, G.J.R. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158.
J Struct Biol X, 4, 2020
|
|
6EP8
 
 | | InhA Y158F mutant in complex with NADH from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Wagner, T, Voegeli, B, Rosenthal, R.G, Stoffel, G, Shima, S, Kiefer, P, Cortina, N, Erb, T.J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | InhA, the enoyl-thioester reductase fromMycobacterium tuberculosisforms a covalent adduct during catalysis.
J. Biol. Chem., 293, 2018
|
|
