8T6K
 
 | |
8T6Q
 
 | |
8W0S
 
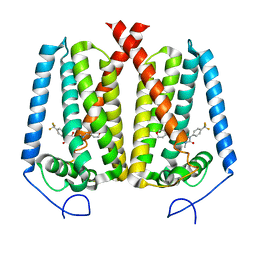 | | Human EBP complexed with compound 3a | | Descriptor: | 1-methyl-8-[(oxan-4-yl)methyl]-3-[4-(trifluoromethyl)phenyl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery and Optimization of Selective Brain-Penetrant EBP Inhibitors that Enhance Oligodendrocyte Formation.
J.Med.Chem., 67, 2024
|
|
8W0R
 
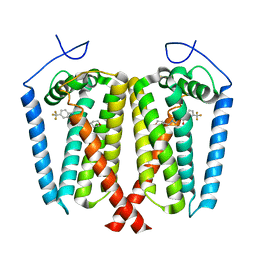 | | Human EBP complexed with compound 1 | | Descriptor: | 1-methyl-1'-[(oxan-4-yl)methyl]-5-(trifluoromethyl)spiro[indole-2,4'-piperidin]-3(1H)-one, 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery and Optimization of Selective Brain-Penetrant EBP Inhibitors that Enhance Oligodendrocyte Formation.
J.Med.Chem., 67, 2024
|
|
8T18
 
 | |
8T15
 
 | |
8T17
 
 | |
8SC7
 
 | |
8VX0
 
 | | CRYSTAL STRUCTURE OF CYP2C9*14 IN COMPLEX WITH LOSARTAN | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Shah, M.B. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural and biophysical analysis of cytochrome P450 2C9*14 and *27 variants in complex with losartan.
J.Inorg.Biochem., 258, 2024
|
|
8VZ7
 
 | |
8SC8
 
 | | Structure of PI3KG in complex with MTX-531 | | Descriptor: | N-[(5P)-2-chloro-5-(4-{[(1R)-1-phenylethyl]amino}quinazolin-6-yl)pyridin-3-yl]methanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whitehead, C.E, Leopold, J. | | Deposit date: | 2023-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Structure of PI3KG in complex with MTX-531
To Be Published
|
|
8PI3
 
 | | Cathepsin S Y132D mutant in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 2024
|
|
8RND
 
 | | Cathepsin S in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin S, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 2024
|
|
8PIX
 
 | | Cryo EM structure of the type 3C polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PJO
 
 | | Cryo EM structure of the type 3D polymorph of alpha-synuclein E46K mutant at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PK2
 
 | | Cryo EM structure of the type 1m polymorph of alpha-synuclein | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8KB0
 
 | | Crystal structure of 01JD-AEAIIVAMV | | Descriptor: | Beta2-microglobulin, MHC class I antigen alpha chain, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8KB1
 
 | | Crystal structure of 11JD | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8QFY
 
 | |
8YXN
 
 | |
8YXK
 
 | |
8OYU
 
 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWV
 
 | | H6 and F2 nanobodies bound to SARS-CoV-2 spike RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2, GLYCEROL, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8RNH
 
 | | Crystal structure of HLA B*18:01 in complex with EEIEITTHF, an 9-mer epitope from Influenza A | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murdolo, L.D, Maddumaage, J, Gras, S. | | Deposit date: | 2024-01-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterisation of novel influenza-derived HLA-B*18:01-restricted epitopes.
Clin Transl Immunology, 13, 2024
|
|
