1G5I
 
 | | CRYSTAL STRUCTURE OF THE ACCESSORY SUBUNIT OF MURINE MITOCHONDRIAL POLYMERASE GAMMA | | Descriptor: | GLYCEROL, MITOCHONDRIAL DNA POLYMERASE ACCESSORY SUBUNIT, SODIUM ION | | Authors: | Carrodeguas, J.A, Theis, K, Bogenhagen, D.F, Kisker, C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer.
Mol.Cell, 7, 2001
|
|
3QBW
 
 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
4OOR
 
 | |
4OPD
 
 | | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase. | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GERANYLGERANYL DIPHOSPHATE | | Authors: | McAndrew, R.P, Kung, Y, Xie, X, Liu, C, Pereira, J.H, Keasling, J.D, Adams, P.D. | | Deposit date: | 2014-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase.
Structure, 22, 2014
|
|
1G3I
 
 | | CRYSTAL STRUCTURE OF THE HSLUV PROTEASE-CHAPERONE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HSLU PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Sousa, M.C, Trame, C.B, Tsuruta, H, Wilbanks, S.M, Reddy, V.S, McKay, D.B. | | Deposit date: | 2000-10-24 | | Release date: | 2000-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Crystal and solution structures of an HslUV protease-chaperone complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1BEA
 
 | | BIFUNCTIONAL HAGEMAN FACTOR/AMYLASE INHIBITOR FROM MAIZE | | Descriptor: | BIFUNCTIONAL AMYLASE/SERINE PROTEASE INHIBITOR | | Authors: | Behnke, C.A, Yee, V.C, Le Trong, I, Pedersen, L.C, Stenkamp, R.E, Kim, S.S, Reeck, G.R, Teller, D.C. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution.
Biochemistry, 37, 1998
|
|
4AFN
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa at 2.3A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, PENTAETHYLENE GLYCOL | | Authors: | Cukier, C.D, Schnell, R, Schneider, G, Lindqvist, Y. | | Deposit date: | 2012-01-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
6EYK
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic ligand NV355 | | Descriptor: | (2~{S})-3-cyclohexyl-2-[(2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[(1~{R},2~{R},3~{S})-3-methyl-2-[(2~{R},3~{S},4~{R},5~{S},6~{R})-3,4,5-tris(oxidanyl)-6-(trifluoromethyl)oxan-2-yl]oxy-cyclohexyl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Zihlmann, P, Preston, R.C, Varga, N, Ernst, B, Maier, T. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Strengthening an Intramolecular Non-Classical Hydrogen Bond to Get in Shape for Binding.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4RAN
 
 | | Aza-acyclic nucleoside phosphonates containing a second phosphonate group as inhibitors of the human, Plasmodium falciparum and vivax 6-oxopurine phosphoribosyltransferases and their pro-drugs as antimalarial agents | | Descriptor: | (2-{[2-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl](3-aminopropyl)amino}ethyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Keough, D.T, Hockova, D, Janeba, Z, Wang, T.-H, Naesens, L, Edstein, M.D, Chavchich, M, Guddat, L.W. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Aza-acyclic Nucleoside Phosphonates Containing a Second Phosphonate Group As Inhibitors of the Human, Plasmodium falciparum and vivax 6-Oxopurine Phosphoribosyltransferases and Their Prodrugs As Antimalarial Agents.
J.Med.Chem., 58, 2015
|
|
1R2I
 
 | | Human Bcl-XL containing a Phe to Leu mutation at position 146 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | O'Neill, J.W, Manion, M.K, Giedt, C.D, Kim, K.M, Zhang, K.Y, Hockenbery, D.M. | | Deposit date: | 2003-09-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bcl-XL mutations suppress cellular sensitivity to antimycin A.
J.Biol.Chem., 279, 2004
|
|
1XN0
 
 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With (R,S)-Rolipram | | Descriptor: | MAGNESIUM ION, ROLIPRAM, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
1R2E
 
 | | Human Bcl-XL containing a Glu to Leu mutation at position 92 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | O'Neill, J.W, Manion, M.K, Giedt, C.D, Kim, K.M, Zhang, K.Y, Hockenbery, D.M. | | Deposit date: | 2003-09-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bcl-XL mutations suppress cellular sensitivity to antimycin A.
J.Biol.Chem., 279, 2004
|
|
3QBX
 
 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to 1,6-anhydro-n-actetylmuramic acid | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
4OYH
 
 | | Structure of Bacillus subtilis MobB | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Kim, D, Choe, J. | | Deposit date: | 2014-02-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Bacillus subtilis MobB
To Be Published
|
|
8E97
 
 | | PYD-106-bound Human GluN1a-GluN2C NMDA receptor in splayed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
1GJB
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
5G37
 
 | | MR structure of the binary mosquito larvicide BinAB at pH 5 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2016-04-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
1AUZ
 
 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3IA2
 
 | | Pseudomonas fluorescens esterase complexed to the R-enantiomer of a sulfonate transition state analog | | Descriptor: | (2R)-butane-2-sulfonate, Arylesterase, GLYCEROL, ... | | Authors: | Schrag, J.D, Kazlauskas, R.J, Jiang, Y, Morley, K. | | Deposit date: | 2009-07-13 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Different active-site loop orientation in serine hydrolases versus acyltransferases.
Chembiochem, 12, 2011
|
|
3GB4
 
 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Cobalt and Dicamba | | Descriptor: | 3,6-dichloro-2-methoxybenzoic acid, COBALT (II) ION, DdmC, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-02-18 | | Release date: | 2009-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
4KBL
 
 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
4GB3
 
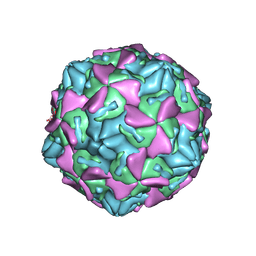 | | Human coxsackievirus B3 strain RD coat protein | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, coat protein 1, ... | | Authors: | Yoder, J.D, Hafenstein, S. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface.
J.Virol., 86, 2012
|
|
1XP0
 
 | | Catalytic Domain Of Human Phosphodiesterase 5A In Complex With Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-07 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
5UZ0
 
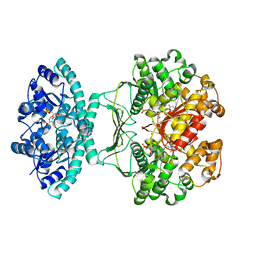 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, MAGNESIUM ION, ... | | Authors: | Atwell, S, Wang, Y, Fales, K.R, Clawson, D, Wang, J. | | Deposit date: | 2017-02-24 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
