3LEW
 
 | |
4RUZ
 
 | |
3SNG
 
 | | X-ray structure of fully glycosylated bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-06-29 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7GKC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-9c80c481-1 (Mpro-P0845) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-{[2-(methylamino)-2-oxoethoxy]methyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GL5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-10fcb19e-1 (Mpro-P1661) | | Descriptor: | (4S)-6-chloro-N-{6-[(methanesulfonyl)amino]isoquinolin-4-yl}-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2Y9Q
 
 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | MAP KINASE-INTERACTING SERINE/THREONINE-PROTEIN KINASE 1, MITOGEN-ACTIVATED PROTEIN KINASE 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Barkai, T, Garai, A, Toeroe, I, Remenyi, A. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
3LO2
 
 | |
3CX4
 
 | |
7GI3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-P0041) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GJX
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-3ccb8ef6-1 (Mpro-P0744) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3D7G
 
 | |
5ZLA
 
 | | Crystal structure of mutant C387A of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase C387A mutant | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5Z9X
 
 | | Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 in complex with an RNA substrate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*GP*CP*CP*CP*AP*UP*UP*AP*G)-3'), SULFATE ION, ... | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Gan, J, Cao, C, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
3LGZ
 
 | | Crystal structure of dehydrosqualene synthase Y129A from S. aureus complexed with presqualene pyrophosphate | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Lin, F.-Y, Liu, Y.-L, Liu, C.-I, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-21 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4DFF
 
 | | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia | | Descriptor: | 8,9-dimethoxy-1-(1,3-thiazol-5-yl)-5,6-dihydroimidazo[5,1-a]isoquinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ho, G.D, Seganish, W.M, Bercovici, A, Tulshian, D, Greenlee, W.J, Van Rijn, R, Hruza, A, Xiao, L, Rindgen, D, Mullins, D, Guzzi, M, Zhang, X, Bleichardt, C, Hodgson, R. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3LJ3
 
 | | PI3-kinase-gamma with a pyrrolopyridine-benzofuran inhibitor | | Descriptor: | (2Z)-4,6-dihydroxy-2-{[1-methyl-4-(4-methylpiperazin-1-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]methylidene}-1-benzofuran-3(2H)-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2010-01-25 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery and optimization of 2-(4-substituted-pyrrolo[2,3-b]pyridin-3-yl)methylene-4-hydroxybenzofuran-3(2H)-ones as potent and selective ATP-competitive inhibitors of the mammalian target of rapamycin (mTOR).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2YFR
 
 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
5V88
 
 | | Structure of DCN1 bound to NAcM-COV | | Descriptor: | Lysozyme,DCN1-like protein 1, N-{2-[({1-[(2R)-pentan-2-yl]piperidin-4-yl}{[3-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]phenyl}propanamide | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
4DNK
 
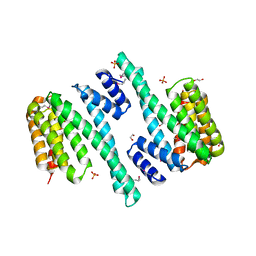 | |
4U0I
 
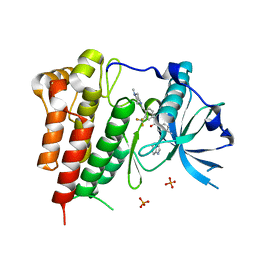 | | Crystal structure of KIT in complex with ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, PHOSPHATE ION | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ponatinib Inhibits Polyclonal Drug-Resistant KIT Oncoproteins and Shows Therapeutic Potential in Heavily Pretreated Gastrointestinal Stromal Tumor (GIST) Patients.
Clin.Cancer Res., 20, 2014
|
|
5VBR
 
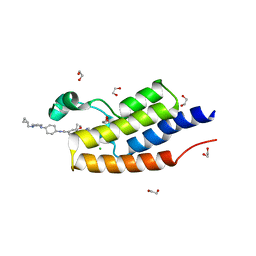 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRDT IN COMPLEX WITH Volasertib | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain testis-specific protein, CHLORIDE ION, ... | | Authors: | EMBER, S.W, ZHU, J.-Y, SCHONBRUNN, E. | | Deposit date: | 2017-03-30 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 2021
|
|
4TVX
 
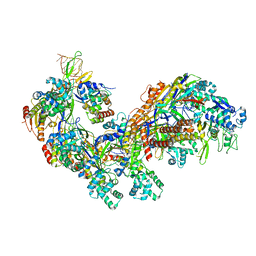 | | Crystal structure of the E. coli CRISPR RNA-guided surveillance complex, Cascade | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Jackson, R.N, Golden, S.M, Carter, J, Wiedenheft, B. | | Deposit date: | 2014-06-28 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli.
Science, 345, 2014
|
|
2YCF
 
 | | Crystal Structure of Checkpoint Kinase 2 in complex with Inhibitor PV1531 | | Descriptor: | (2E)-N-hydroxy-2-[1-(4-{[(4-{(1E)-1-[2-(N'-hydroxycarbamimidoyl)hydrazinylidene]ethyl}phenyl)carbamoyl]amino}phenyl)ethylidene]hydrazinecarboximidamide, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-14 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
7GO3
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1198162455 | | Descriptor: | 1-(1-ethyl-1H-pyrazol-3-yl)-N-methylmethanamine, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
