3FKL
 
 | | P38 kinase crystal structure in complex with RO9552 | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-[8-methyl-7-oxo-2-(tetrahydro-2H-pyran-4-ylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl]benzamide | | Authors: | Kuglstatter, A, Ghate, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Binding Pocket Volume: Potential Applications towards Ligand Design and Selectivity
To be Published
|
|
3FKO
 
 | | P38 kinase crystal structure in complex with RO3668 | | Descriptor: | 3-(2-chlorophenyl)-6-(2-fluorophenoxy)-2H-indazole, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Knapp, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Binding Pocket Volume: Potential Applications towards Ligand Design and Selectivity
To be Published
|
|
2EWA
 
 | | Dual binding mode of pyridinylimidazole to MAP kinase p38 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | Delarbre, L, Pouzieux, S, Guilloteau, J.-P, Michot, N. | | Deposit date: | 2005-11-02 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NMR characterization of kinase p38 dynamics in free and ligand-bound forms.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
5FLV
 
 | | Crystal structure of NKX2-5 and TBX5 bound to the Nppa promoter region | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP *GP*TP*GP*AP*GP*AP*AP*G)-3', 5'-D(*TP*CP*TP*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP *TP*GP*AP*AP*GP*TP*GP*G)-3', HOMEOBOX PROTEIN NKX-2.5, ... | | Authors: | Stirnimann, C.U, Glatt, S, Mueller, C.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis.
Cell(Cambridge,Mass.), 164, 2016
|
|
8TO0
 
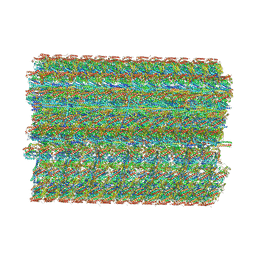 | | 48-nm repeating structure of doublets from mouse sperm flagella | | Descriptor: | Cilia- and flagella- associated protein 210, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Chen, Z, Shiozak, M, Hass, K.M, Skinner, W, Zhao, S, Guo, C, Polacco, B.J, Yu, Z, Krogan, N.J, Kaake, R.M, Vale, R.D, Agard, D.A. | | Deposit date: | 2023-08-02 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | De novo protein identification in mammalian sperm using in situ cryoelectron tomography and AlphaFold2 docking.
Cell, 186, 2023
|
|
5FYQ
 
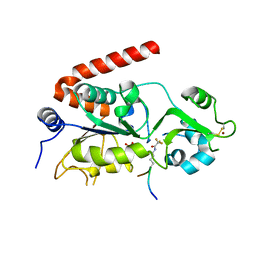 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
7PSX
 
 | | Structure of HOXB13 bound to hydroxymethylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*5HCP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2021-09-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of HOXB13 bound to hydroxymethylated DNA
To Be Published
|
|
2LGC
 
 | |
4NEJ
 
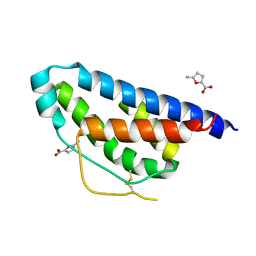 | | Small molecular fragment bound to crystal contact interface of Interleukin-2 | | Descriptor: | 5-methylfuran-2-carboxylic acid, Interleukin-2 | | Authors: | Brenke, R, Jehle, S, Vajda, S, Allen, K.N, Kozakov, D. | | Deposit date: | 2013-10-29 | | Release date: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Small molecular fragments bound to binding energy hot-spot in crystal contact interface of Interleukin-2
To be Published
|
|
7Q3O
 
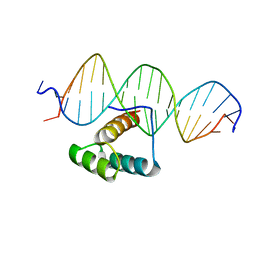 | |
8IFG
 
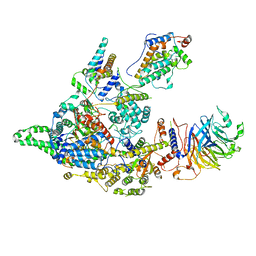 | | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Cph2, ... | | Authors: | Zhang, H.Q, Wang, X, Wang, Y.N, Liu, S.M, Zhang, Y, Xu, K, Ji, L.T, Kornberg, R.D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Class I histone deacetylase complex: Structure and functional correlates.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8A4F
 
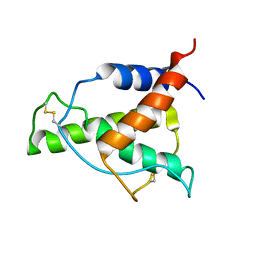 | | Human Interleukin-4 mutant - C3T-IL4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Mueller, T.D, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2022-06-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
7QHS
 
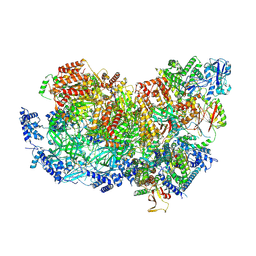 | | S. cerevisiae CMGE nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
5VHM
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
6DKS
 
 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | Deposit date: | 2018-05-30 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
5CXR
 
 | |
7C9Q
 
 | | Crystal structure of Human liver fructose-1,6-bisphoaphatase complex with Mg2+ and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION | | Authors: | Huang, Y, Li, R, Wan, J. | | Deposit date: | 2020-06-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Crystal structure of Human liver fructose-1,6-bisphoaphatase complex with Mg2+ and AMP
To Be Published
|
|
4NEM
 
 | | Small molecular fragment bound to crystal contact interface of Interleukin-2 | | Descriptor: | 5-[(2,3-dichlorophenoxy)methyl]furan-2-carboxylic acid, Interleukin-2 | | Authors: | Jehle, S, Brenke, R, Vajda, S, Allen, K.N, Kozakov, D. | | Deposit date: | 2013-10-29 | | Release date: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Small molecular fragments bound to binding energy hot-spot in crystal contact interface of Interleukin-2
To be Published
|
|
7CWE
 
 | | Human Fructose-1,6-bisphosphatase 1 in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1, MAGNESIUM ION | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 in APO R-state
To Be Published
|
|
3A4S
 
 | | The crystal structure of the SLD2:Ubc9 complex | | Descriptor: | NFATC2-interacting protein, SUMO-conjugating enzyme UBC9 | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
7CVH
 
 | | Human Fructose-1,6-bisphosphatase 1 in complex with geranylgeranyl diphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structural basis for GGPP activation on the enzymatic activities FBP1
To Be Published
|
|
7DNO
 
 | | Characterization of Peptide Ligands Against WDR5 Isolated Using Phage Display Technique | | Descriptor: | CYS-ARG-THR-LEU-PRO-PHE, WD repeat-containing protein 5 | | Authors: | Cao, J, Cao, D, Xiong, B, Li, Y, Fan, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5.
Molecules, 26, 2021
|
|
5VHO
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
8UII
 
 | |
8UI7
 
 | |
