5F9H
 
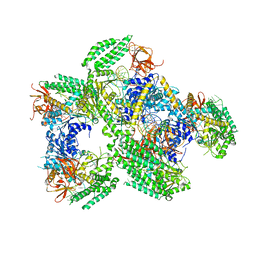 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer 5' triphosphate hairpin RNA | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3LRR
 
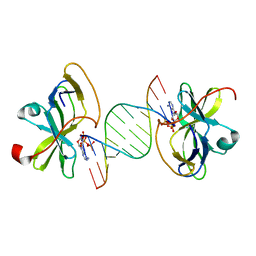 | | Crystal structure of human RIG-I CTD bound to a 12 bp AU rich 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(ATP)P*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*U)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
3LRN
 
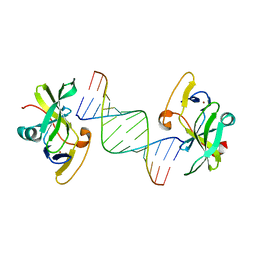 | | Crystal structure of human RIG-I CTD bound to a 14 bp GC 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
2QFD
 
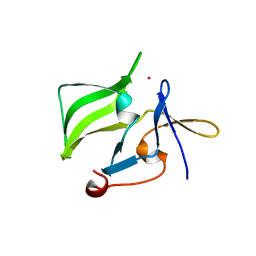 | | Crystal structure of the regulatory domain of human RIG-I with bound Hg | | Descriptor: | MERCURY (II) ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Cui, S, Lammens, A, Lammens, K, Hopfner, K.P. | | Deposit date: | 2007-06-27 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I.
Mol.Cell, 29, 2008
|
|
4AY2
 
 | | Capturing 5' tri-phosphorylated RNA duplex by RIG-I | | Descriptor: | 5'-R-PPP(GP*GP*CP*GP*CP*GP*GP*CP*UP*UP*CP*GP*GP*CP *CP*GP*CP*GP*CP*C)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2012-06-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Visualizing the Determinants of Viral RNA Recognition by Innate Immune Sensor Rig-I.
Structure, 20, 2012
|
|
3ZD6
 
 | | Snapshot 1 of RIG-I scanning on RNA duplex | | Descriptor: | PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, RNA DUPLEX, ZINC ION | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2012-11-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defining the Functional Determinants for RNA Surveillance by Rig-I.
Embo Rep., 14, 2013
|
|
3ZD7
 
 | | Snapshot 3 of RIG-I scanning on RNA duplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2012-11-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Defining the Functional Determinants for RNA Surveillance by Rig-I.
Embo Rep., 14, 2013
|
|
2QFB
 
 | | Crystal structure of the regulatory domain of human RIG-I with bound Zn | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, ZINC ION | | Authors: | Cui, S, Lammens, A, Lammens, K, Hopfner, K.P. | | Deposit date: | 2007-06-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I.
Mol.Cell, 29, 2008
|
|
8FKY
 
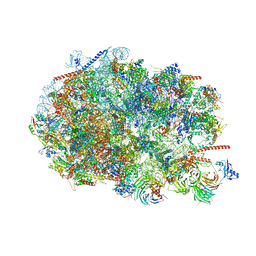 | |
8FKT
 
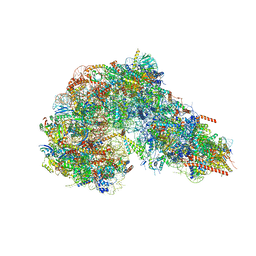 | |
8FKV
 
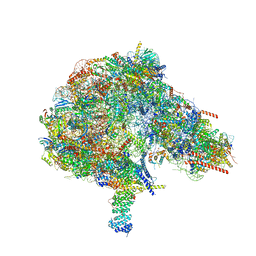 | |
8FKU
 
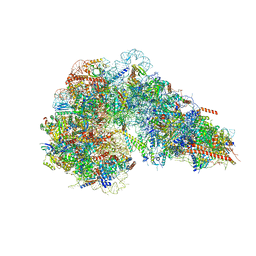 | |
8FKX
 
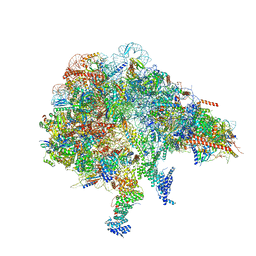 | |
8FKW
 
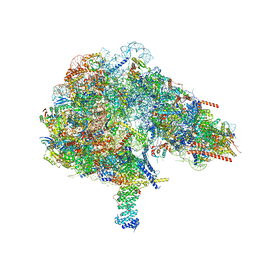 | |
7MK1
 
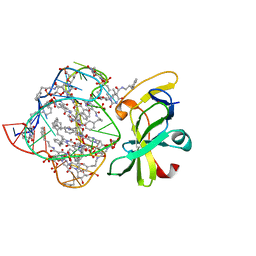 | | Structure of a protein-modified aptamer complex | | Descriptor: | Antiviral innate immune response receptor RIG-I, DNA (41-MER), MAGNESIUM ION, ... | | Authors: | Ren, X, Pyle, A.M. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving A RIG-I Antagonist: A Modified DNA Aptamer Mimics Viral RNA.
J.Mol.Biol., 433, 2021
|
|
3TBK
 
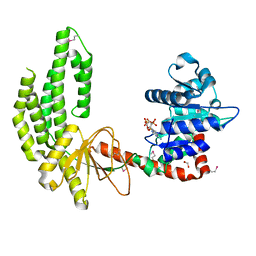 | | Mouse RIG-I ATPase Domain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RIG-I Helicase Domain | | Authors: | Civril, F, Bennett, M.D, Hopfner, K.-P. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling.
Embo Rep., 12, 2011
|
|
8SD0
 
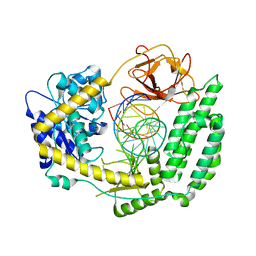 | |
7BAH
 
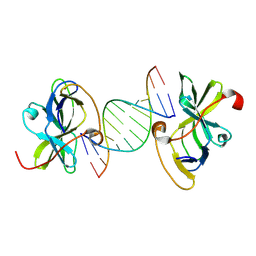 | | Structure of RIG-I CTD bound to OH-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A conserved isoleucine in the RNA sensor RIG-I controls immune tolerance to mitochondrial RNA
To Be Published
|
|
7JL3
 
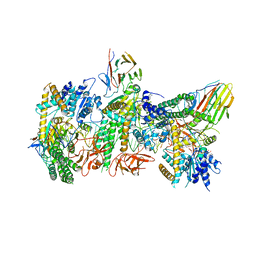 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL1
 
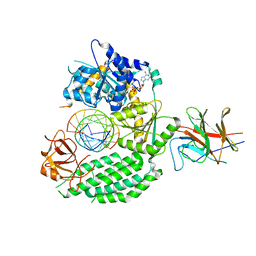 | | Cryo-EM structure of RIG-I:dsRNA in complex with RIPLET PrySpry domain (monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7BAI
 
 | |
8DVR
 
 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+AMPPNP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, GUANOSINE-5'-TRIPHOSPHATE, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVS
 
 | | Cryo-EM structure of RIG-I bound to the end of OHSLR30 (+ATP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, MAGNESIUM ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVU
 
 | |
3NCU
 
 | |
