5UZI
 
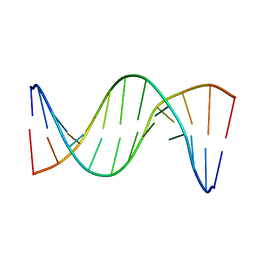 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNAm1A16 structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*(M1A)P*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
4FLM
 
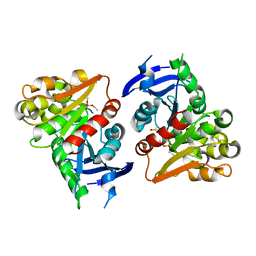 | | S-formylglutathione Hydrolase W197I Variant containing Copper | | Descriptor: | COPPER (II) ION, S-formylglutathione hydrolase | | Authors: | Legler, P.M, Millard, C.B. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A role for His-160 in peroxide inhibition of S. cerevisiae S-formylglutathione hydrolase: Evidence for an oxidation sensitive motif.
Arch.Biochem.Biophys., 528, 2012
|
|
3NCB
 
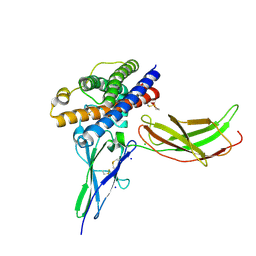 | | A mutant human Prolactin receptor antagonist H180A in complex with the extracellular domain of the human prolactin receptor | | Descriptor: | CARBONATE ION, CHLORIDE ION, Prolactin, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
4FP1
 
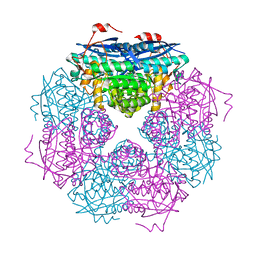 | | P. putida mandelate racemase co-crystallized with 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl) propionic acid | | Descriptor: | 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl)propanoic acid, MAGNESIUM ION, Mandelate racemase | | Authors: | Lietzan, A.D, St.Maurice, M. | | Deposit date: | 2012-06-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Potent inhibition of mandelate racemase by a fluorinated substrate-product analogue with a novel binding mode.
Biochemistry, 53, 2014
|
|
5IEY
 
 | | Crystal structure of a CDK inhibitor bound to CDK2 | | Descriptor: | 4-[(4-{[(2R,3R)-3-hydroxybutan-2-yl]amino}pyrimidin-2-yl)amino]benzene-1-sulfonamide, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
3D81
 
 | | Sir2-S-alkylamidate complex crystal structure | | Descriptor: | NAD-dependent deacetylase, S-alkylamidate intermediate, ZINC ION | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
5CC9
 
 | |
5IG9
 
 | |
5C3F
 
 | | Crystal structure of Mcl-1 bound to BID-MM | | Descriptor: | BID-MM, GLYCEROL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
6FML
 
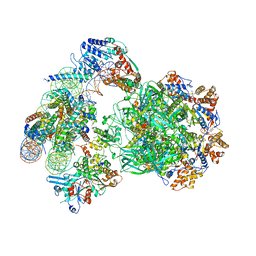 | | CryoEM Structure INO80core Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin related protein 5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
3NGS
 
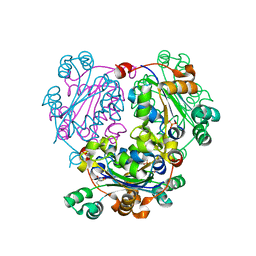 | | Structure of Leishmania nucleoside diphosphate kinase b with ordered nucleotide-binding loop | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Nucleoside diphosphate kinase, PHOSPHATE ION | | Authors: | Trindade, D.M, Sousa, T.A.C.B, Tonoli, C.C.C, Santos, C.R, Arni, R.K, Ward, R.J, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-06-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
3NCF
 
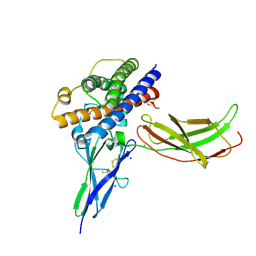 | | A mutant human Prolactin receptor antagonist H30A in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
6E8S
 
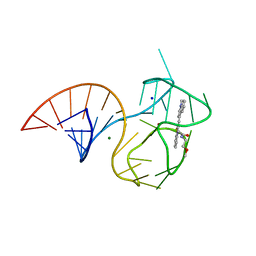 | | Structure of the iMango-III aptamer bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.
Nat. Chem. Biol., 15, 2019
|
|
3CGU
 
 | |
5USH
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv66 | | Descriptor: | Fab vv66 heavy chain, Fab vv66 light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
6E9A
 
 | | HIV-1 WILD TYPE PROTEASE WITH GRL-034-17A, (3aS, 5R, 6aR)-2-OXOHEXAHYD CYCLOPENTA[D]-5-OXAZOLYL URETHANE WITH A BICYCLIC OXAZOLIDINONE SCAFF AS THE P2 LIGAND | | Descriptor: | (3aS,5R,6aR)-2-oxohexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
5INF
 
 | | Structural basis for acyl-CoA carboxylase-mediated assembly of unusual polyketide synthase extender units incorporated into the stambomycin antibiotics | | Descriptor: | Carboxyl transferase, HEXANOYL-COENZYME A | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Withall, D.M, Song, L, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
7MZY
 
 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-986 | | Descriptor: | ACETATE ION, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Sowaileh, M, Miller, D.J, Rossi, P, Myasnikov, A.G, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
2AJM
 
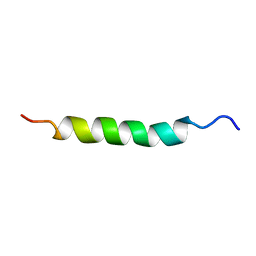 | | NMR structure of the in-plane membrane anchor domain [1-28] of the monotopic NonStructural Protein 5A (NS5A) from the Bovine Viral Diarrhea Virus (BVDV) | | Descriptor: | Nonstructural protein 5A | | Authors: | Sapay, N, Montserret, R, Chipot, C, Brass, V, Moradpour, D, Deleage, G, Penin, F. | | Deposit date: | 2005-08-02 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus.
Biochemistry, 45, 2006
|
|
6EJD
 
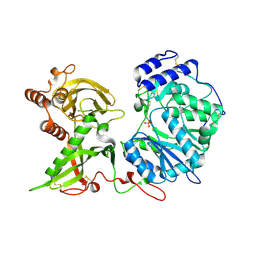 | |
3H8N
 
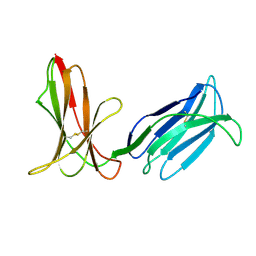 | | Crystal Structure Analysis of KIR2DS4 | | Descriptor: | Killer cell immunoglobulin-like receptor 2DS4 | | Authors: | Graef, T, Bushnell, D.A, Parham, P. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C.
J.Exp.Med., 206, 2009
|
|
5IQ3
 
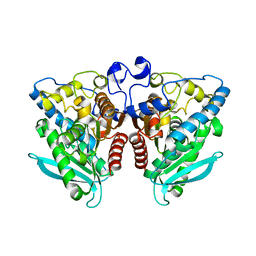 | |
5V4I
 
 | | Osmium(II)(cymene)(chlorido)2-lysozyme adduct with one binding site | | Descriptor: | Lysozyme C, SODIUM ION, dichloro[(1,2,3,4,5,6-eta)-3-methyl-6-(propan-2-yl)benzene-1,2,4,5-tetrayl]osmium | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The metalation of hen egg white lysozyme impacts protein stability as shown by ion mobility mass spectrometry, differential scanning calorimetry, and X-ray crystallography.
Chem. Commun. (Camb.), 53, 2017
|
|
3K9Z
 
 | | Rational Design of a Structural and Functional Nitric Oxide Reductase | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yeung, N, Lin, Y.-W, Gao, Y.-G, Zhao, X, Russell, B.S, Lei, L, Miner, K.D, Robinson, H, Lu, Y. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of a structural and functional nitric oxide reductase.
Nature, 462, 2009
|
|
5BZY
 
 | |
