8YHF
 
 | | The Crystal Structure of the Type I TGF beta receptor from Biortus. | | Descriptor: | 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, TGF-beta receptor type-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Type I TGF beta receptor from Biortus.
To Be Published
|
|
8USO
 
 | | Full-length human CaMKII delta holoenzyme | | Descriptor: | MALONATE ION, calcium/calmodulin-dependent protein kinase | | Authors: | Ozden, C, Abromson, N.L, Tomchick, D.R, Stratton, M.M, Garman, S.C. | | Deposit date: | 2023-10-28 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length human CaMKII delta holoenzyme
To be published
|
|
1YM7
 
 | | G Protein-Coupled Receptor Kinase 2 (GRK2) | | Descriptor: | Beta-adrenergic receptor kinase 1 | | Authors: | Lodowski, D.T, Barnhill, J.F, Pyskadlo, R.M, Ghirlando, R, Sterne-Marr, R, Tesmer, J.J.G. | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The role of Gbetagamma and domain interfaces in the activation of G protein-coupled receptor kinase 2
Biochemistry, 44, 2005
|
|
8TXY
 
 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UAK
 
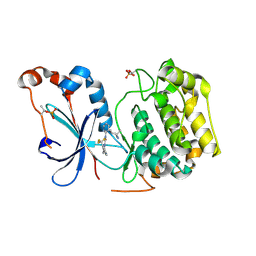 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with Darovasertib (NVP-LXS196) at 2.82-A resolution | | Descriptor: | (6M)-3-amino-N-[3-(4-amino-4-methylpiperidin-1-yl)pyridin-2-yl]-6-[3-(trifluoromethyl)pyridin-2-yl]pyrazine-2-carboxamide, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8U7H
 
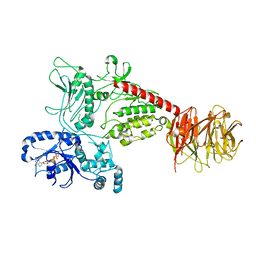 | | Cryo-EM structure of LRRK2 bound to type I inhibitor GNE-7915 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, [4-[[4-(ethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-2-fluoranyl-5-methoxy-phenyl]-morpholin-4-yl-methanone, non-specific serine/threonine protein kinase | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
5LI9
 
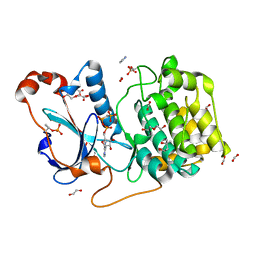 | | Structure of a nucleotide-bound form of PKCiota core kinase domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
5LQF
 
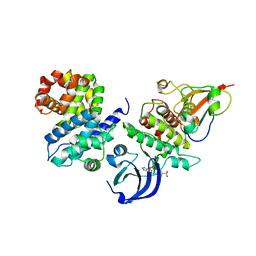 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
5LMK
 
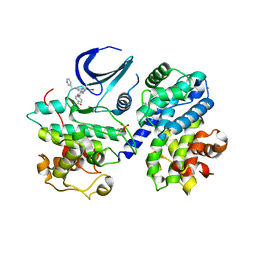 | | Structure of phopsho-CDK2-cyclin A in complex with an ATP-competitive inhibitor | | Descriptor: | 4-[4-[3-bromanyl-7-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-5-yl]phenyl]benzamide, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Explicit treatment of active-site waters enhances quantum mechanical/implicit solvent scoring: Inhibition of CDK2 by new pyrazolo[1,5-a]pyrimidines.
Eur J Med Chem, 126, 2016
|
|
1UNL
 
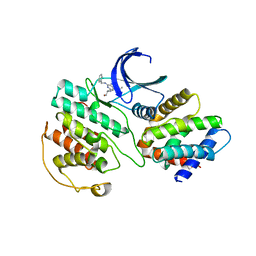 | | Structural mechanism for the inhibition of CD5-p25 from the roscovitine, aloisine and indirubin. | | Descriptor: | CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1, R-ROSCOVITINE | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNH
 
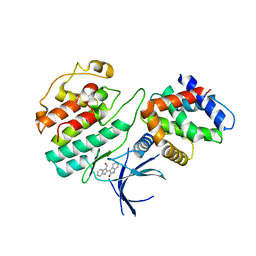 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNG
 
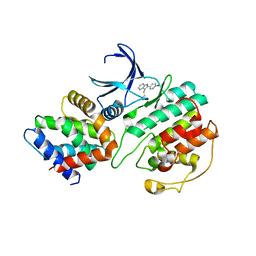 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | 6-PHENYL[5H]PYRROLO[2,3-B]PYRAZINE, CELL DIVISION PROTEIN KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
5M5A
 
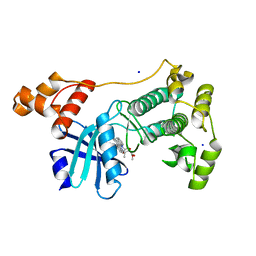 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, K-252A, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-10-21 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5LXM
 
 | |
1XJD
 
 | |
9C82
 
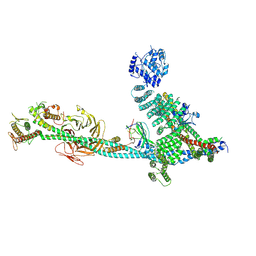 | | Structure of human ULK1C:PI3KC3-C1 supercomplex | | Descriptor: | Beclin 1-associated autophagy-related key regulator, Beclin-1, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
1Y8G
 
 | |
1XO2
 
 | | Crystal structure of a human cyclin-dependent kinase 6 complex with a flavonol inhibitor, fisetin | | Descriptor: | 3,7,3',4'-TETRAHYDROXYFLAVONE, Cell division protein kinase 6, Cyclin | | Authors: | Lu, H.S, Chang, D.J, Baratte, B, Meijer, L, Schulze-Gahmen, U. | | Deposit date: | 2004-10-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Human Cyclin-Dependent Kinase 6 Complex with a Flavonol Inhibitor, Fisetin.
J.Med.Chem., 48, 2005
|
|
1ZMW
 
 | |
1ZMU
 
 | |
1ZMV
 
 | |
3C4Y
 
 | |
3C51
 
 | |
3C4X
 
 | |
3D0E
 
 | | Crystal structure of human Akt2 in complex with GSK690693 | | Descriptor: | 4-{2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-[(3S)-piperidin-3-ylmethoxy]-1H-imidazo[4,5-c]pyridin-4-yl}-2-methylbut-3-yn-2-ol, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Smallwood, A. | | Deposit date: | 2008-05-01 | | Release date: | 2008-10-21 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H-imidazo[4,5-c]pyridin-4-yl)-2-methyl-3-butyn-2-ol (GSK690693), a novel inhibitor of AKT kinase.
J.Med.Chem., 51, 2008
|
|
