1EP0
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
5CI5
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein family 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose
To be published
|
|
1BIO
 
 | | HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL, ISATOIC ANHYDRIDE | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
6IYV
 
 | | Crystal sturucture of L,D-transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with ertapenem adduct | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhao, F, Li, D.F, Wang, D.C. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1-beta-methyl group confers a lower affinity of l,d-transpeptidase LdtMt2for ertapenem than for imipenem.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
5CKS
 
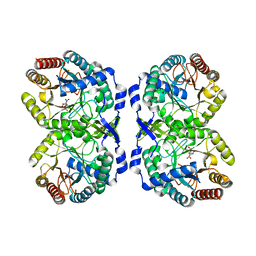 | | DAHP (3-deoxy-D-arabinoheptulosonate-7-phosphate) Synthase in complex with DAHP Oxime. | | Descriptor: | DAHP Oxime, Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive, ... | | Authors: | Berti, P, Junop, M, Balachandran, N. | | Deposit date: | 2015-07-15 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1181 Å) | | Cite: | Potent Inhibition of 3-Deoxy-d-arabinoheptulosonate-7-phosphate (DAHP) Synthase by DAHP Oxime, a Phosphate Group Mimic.
Biochemistry, 55, 2016
|
|
7KJM
 
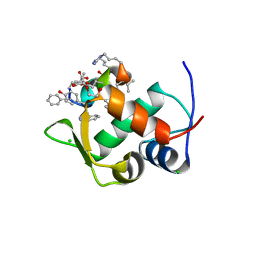 | |
1DN4
 
 | | SOLVATION OF THE LEFT-HANDED HEXAMER D(5BRC-G-5BRC-G-5BRC-G) IN CRYSTALS GROWN AT TWO TEMPERATURES | | Descriptor: | DNA (5'-D(*(CBR)P*GP*(CBR)P*GP*(CBR)P*G)-3') | | Authors: | Chevrier, B, Dock, A.C, Hartmann, B, Leng, M, Moras, D, Thuong, M.T, Westhof, E. | | Deposit date: | 1986-12-01 | | Release date: | 1987-04-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5 BrC-G) in crystals grown at two temperatures.
J.Mol.Biol., 188, 1986
|
|
2XW9
 
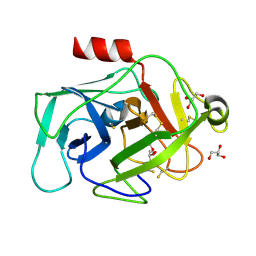 | | Crystal Structure of Complement Factor D mutant S183A | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
1MVS
 
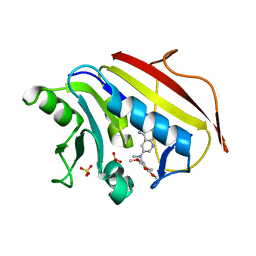 | | Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',4',5'-TRIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate Reductase, SULFATE ION | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W.A, Gangjee, A. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of two polymorphic forms of a pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolate binary complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1M7L
 
 | | Solution Structure of the Coiled-Coil Trimerization Domain from Lung Surfactant Protein D | | Descriptor: | Pulmonary surfactant-associated protein D | | Authors: | Kovacs, H, O'Donoghue, S.I, Hoppe, H.-J, Comfort, D, Reid, K.B.M, Campbell, I.D, Nilges, M. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the coiled-coil trimerization domain from lung surfactant protein D
J.BIOMOL.NMR, 24, 2002
|
|
5TSQ
 
 | | Crystal structure of IUnH from Leishmania braziliensis in complex with D-Ribose | | Descriptor: | CALCIUM ION, IUnH, beta-D-ribofuranose | | Authors: | Bachega, J.F.R, Timmers, L.F.S.M, Dalberto, P.F, Martinelli, L, Pinto, A.F.M, Basso, L.A, Norberto de Souza, O, Santos, D.S. | | Deposit date: | 2016-10-31 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of IUnH from Leishmania braziliensis in complex with D-Ribose
To Be Published
|
|
1DN5
 
 | | SOLVATION OF THE LEFT-HANDED HEXAMER D(5BRC-G-5BRC-G-5BRC-G) IN CRYSTALS GROWN AT TWO TEMPERATURES | | Descriptor: | DNA (5'-D(*(CBR)P*GP*(CBR)P*GP*(CBR)P*G)-3') | | Authors: | Chevrier, B, Dock, A.C, Hartmann, B, Leng, M, Moras, D, Thuong, M.T, Westhof, E. | | Deposit date: | 1986-12-01 | | Release date: | 1987-04-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5 BrC-G) in crystals grown at two temperatures.
J.Mol.Biol., 188, 1986
|
|
1DA0
 
 | | DNA-DRUG INTERACTIONS: THE CRYSTAL STRUCTURE OF D(CGATCG) COMPLEXED WITH DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Moore, M.H, Hunter, W.N, Langlois D'Estaintot, B, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA-drug interactions. The crystal structure of d(CGATCG) complexed with daunomycin.
J.Mol.Biol., 206, 1989
|
|
1LY4
 
 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4HU2
 
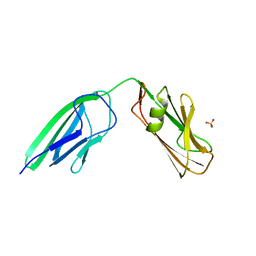 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain A and B | | Descriptor: | PROBABLE CONSERVED LIPOPROTEIN LPPS, SULFATE ION | | Authors: | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1DB1
 
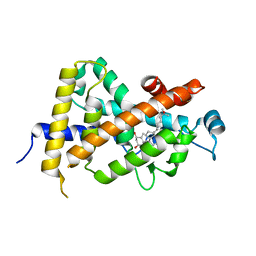 | | CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D NUCLEAR RECEPTOR | | Authors: | Rochel, N, Wurtz, J.M, Mitschler, A, Klaholz, B, Moras, D. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand.
Mol.Cell, 5, 2000
|
|
9FGC
 
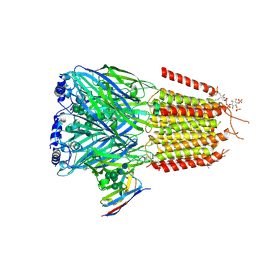 | |
6IYW
 
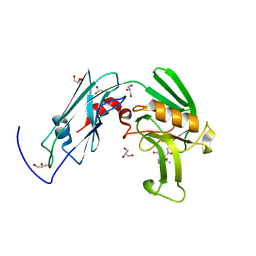 | | Crystal sturucture of L,D-transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with Imipenem adduct | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, GLYCEROL, L,D-transpeptidase 2 | | Authors: | Li, D.F, Zhao, F, Wang, D.C. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1-beta-methyl group confers a lower affinity of l,d-transpeptidase LdtMt2 for ertapenem than for imipenem.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
1HMA
 
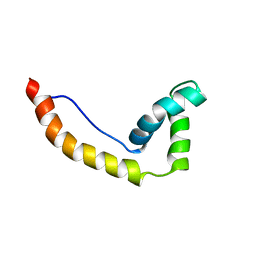 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | Descriptor: | HMG-D | | Authors: | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | Deposit date: | 1994-05-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
1IHH
 
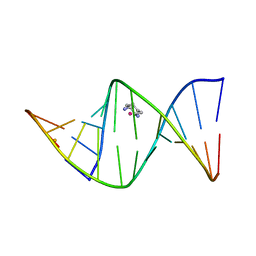 | | 2.4 ANGSTROM CRYSTAL STRUCTURE OF AN OXALIPLATIN 1,2-D(GPG) INTRASTRAND CROSS-LINK IN A DNA DODECAMER DUPLEX | | Descriptor: | 1R,2R-DIAMINOCYCLOHEXANE, 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', ... | | Authors: | Spingler, B, Whittington, D.A, Lippard, S.J. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex.
Inorg.Chem., 40, 2001
|
|
3IKP
 
 | |
5NRI
 
 | |
4P7Q
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3PXK
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH Pyrrolo[2,3-d]thiazole | | Descriptor: | 6-(4,4-dimethylpent-2-ynyl)-4~{H}-pyrrolo[2,3-d][1,3]thiazole, PTK2 protein, SULFATE ION | | Authors: | Koolman, H, Heinrich, T, Musil, D. | | Deposit date: | 2010-12-10 | | Release date: | 2011-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Co-crystal Structures of FAK with an Unprecedented Pyrrolo[2,3-d]thiazole
To be Published
|
|
4NAP
 
 | | Crystal structure of a trap periplasmic solute binding protein from Desulfovibrio alaskensis G20 (DDE_0634), target EFI-510102, with bound d-tryptophan | | Descriptor: | D-TRYPTOPHAN, Extracellular solute-binding protein, family 7 | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
