8F1G
 
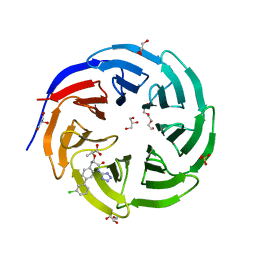 | | Crystal structure of human WDR5 in complex with compound WM662 | | Descriptor: | (2S)-2-({(2S)-3-(3'-chloro[1,1'-biphenyl]-4-yl)-1-oxo-1-[(1H-tetrazol-5-yl)amino]propan-2-yl}oxy)propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, H. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of WDR5-MYC Interaction.
Acs Chem.Biol., 18, 2023
|
|
8EQ8
 
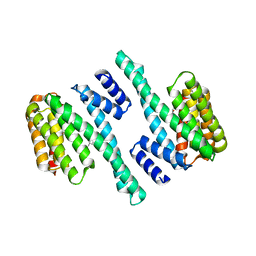 | |
8EQH
 
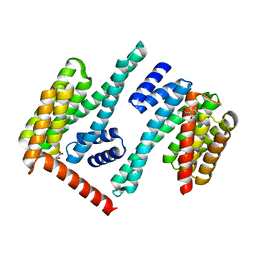 | |
8F0W
 
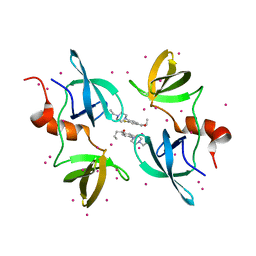 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
4OGI
 
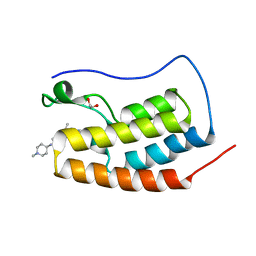 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor BI-2536 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Jose, B, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Dual kinase-bromodomain inhibitors for rationally designed polypharmacology.
Nat.Chem.Biol., 10, 2014
|
|
4OJN
 
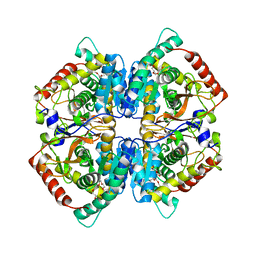 | | Crystal structure of human muscle L-lactate dehydrogenase | | Descriptor: | GLYCEROL, L-lactate dehydrogenase A chain, PENTAETHYLENE GLYCOL | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ZZR
 
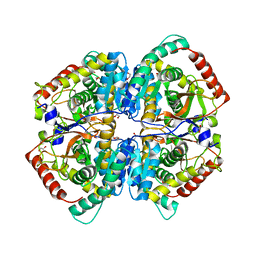 | | The Crystal Structure of human LDHA from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-lactate dehydrogenase A chain | | Authors: | Wang, F, Lin, D, Cheng, W, Bao, X, Zhu, B, Shang, H. | | Deposit date: | 2020-08-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of human LDHA from Biortus
To Be Published
|
|
4OL0
 
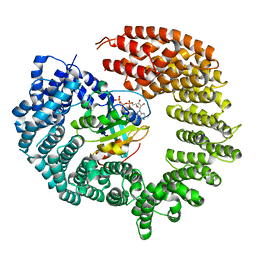 | | Crystal structure of transportin-SR2, a karyopherin involved in human disease, in complex with Ran | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tsirkone, V.G, Strelkov, S.V. | | Deposit date: | 2014-01-23 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of transportin SR2, a karyopherin involved in human disease, in complex with Ran.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7AAD
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
4O76
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with TG101209 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O77
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB 202190 | | Descriptor: | 4-[4-(4-fluorophenyl)-5-(pyridin-4-yl)-1H-imidazol-2-yl]phenol, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O71
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with FLAVOPIRIDOL | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
6ZMD
 
 | | Crystal structure of HYPE covalently tethered to BiP bound to AMP-PNP | | Descriptor: | Endoplasmic reticulum chaperone BiP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fauser, J, Gulen, B, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Specificity of AMPylation of the human chaperone BiP is mediated by TPR motifs of FICD.
Nat Commun, 12, 2021
|
|
4O75
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with FOSTAMATINIB | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [6-({5-fluoro-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}amino)-2,2-dimethyl-3-oxo-2,3-dihydro-4H-pyrido[3,2-b][1,4]oxazin-4-yl]methyl dihydrogen phosphate | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
7A5Y
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with Rp-dGTP-alphaS (T8T) and Mg | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Purkiss, A, Taylor, I.A. | | Deposit date: | 2020-08-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Probing the Catalytic Mechanism and Inhibition of SAMHD1 Using the Differential Properties of R p - and S p -dNTP alpha S Diastereomers.
Biochemistry, 60, 2021
|
|
6ZXO
 
 | |
4OKN
 
 | | Crystal structure of human muscle L-lactate dehydrogenase, ternary complex with NADH and oxalate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, KANAMYCIN A, L-lactate dehydrogenase A chain, ... | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8EYK
 
 | |
7ABT
 
 | |
8EYI
 
 | |
4OGJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor TG-101348 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide | | Authors: | Filippakopoulos, P, Picaud, S, Jose, B, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dual kinase-bromodomain inhibitors for rationally designed polypharmacology.
Nat.Chem.Biol., 10, 2014
|
|
6ZWK
 
 | | Crystal structure of the phosphorylated C-terminal tail of histone H2AX in complex with a specific nanobody (C6 gammaXbody) | | Descriptor: | CHLORIDE ION, Histone H2AX, SODIUM ION, ... | | Authors: | McEwen, A.G, Moeglin, E, Desplancq, D, Weiss, E, Poterszman, A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Novel Nanobody Precisely Visualizes Phosphorylated Histone H2AX in Living Cancer Cells under Drug-Induced Replication Stress.
Cancers (Basel), 13, 2021
|
|
6ZUE
 
 | |
4PCE
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with compound B13 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-2-ethyl-1,5,6,7-tetrahydro-4H-indol-4-one, Bromodomain-containing protein 4 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4PJ1
 
 | | Crystal structure of the human mitochondrial chaperonin symmetrical 'football' complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Frolow, F, Azem, A, Nisemblat, S. | | Deposit date: | 2014-05-10 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the human mitochondrial chaperonin symmetrical football complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
