8ESA
 
 | |
8ERL
 
 | |
8EYW
 
 | | Beetroot dimer bound to ThT | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, POTASSIUM ION, RNA (49-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
6MY0
 
 | |
1MVP
 
 | |
8EYU
 
 | | Structure of Beetroot dimer bound to DFAME | | Descriptor: | POTASSIUM ION, RNA (49-MER), methyl (2E)-3-{(4Z)-4-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-1-methyl-5-oxo-4,5-dihydro-1H-imidazol-2-yl}prop-2-enoate | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
1N27
 
 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
8EYV
 
 | | Structure of Beetroot dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (45-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8F0N
 
 | | Wobble Beetroot (A16U-U38G) dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (49-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-11-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8ESM
 
 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, {(1r,4r)-4-[4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl]cyclohexyl}acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
8ETM
 
 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(1S,4r)-4-{4-[(4S)-2-({[4-(trifluoromethoxy)phenyl]methyl}carbamoyl)imidazo[1,2-a]pyridin-6-yl]phenyl}cyclohexyl]acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
8ESK
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, resting-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
6MWH
 
 | | LasR LBD:BB0020 complex | | Descriptor: | 2-(3-bromophenoxy)-N-[(1S,2S,3R,5S)-2-hydroxybicyclo[3.1.0]hexan-3-yl]acetamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-29 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
8F6Y
 
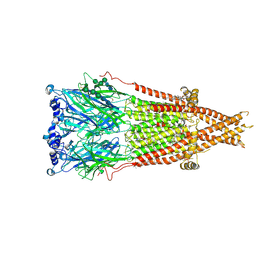 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with etomidate, desensitized-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F6Z
 
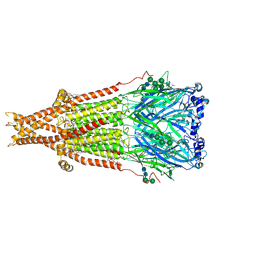 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with succinylcholine, desensitized-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2,2'-[(1,4-DIOXOBUTANE-1,4-DIYL)BIS(OXY)]BIS(N,N,N-TRIMETHYLETHANAMINIUM), 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
6MXX
 
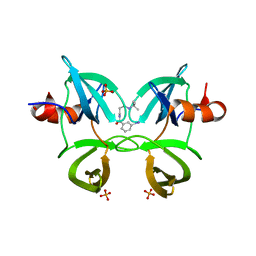 | | Structure of 53BP1 tandem Tudor domains in complex with small molecule UNC2991 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-iodobenzamide, PHOSPHATE ION, ... | | Authors: | Cui, G, Botuyan, M.V, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
8F2S
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, pore-blocked state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
6MSO
 
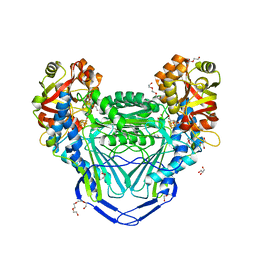 | | Crystal structure of mitochondrial fumarate hydratase from Leishmania major in a complex with inhibitor thiomalate | | Descriptor: | (2S)-2-sulfanylbutanedioic acid, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Feliciano, P.R, Drennan, C.L, Nonato, M.C. | | Deposit date: | 2018-10-17 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Crystal Structures of Fumarate Hydratases from Leishmania major in a Complex with Inhibitor 2-Thiomalate.
ACS Chem. Biol., 14, 2019
|
|
8F5F
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | (2P)-2-[(4P)-4-{6-[(1-ethylcyclopropyl)methoxy]pyridin-3-yl}-1,3-thiazol-2-yl]benzoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
8F5J
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | 3-chloro-5-fluorothieno[3,2-b]thiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
8F5S
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | (2M)-2-[2-(4-methylphenyl)-1,3-thiazol-4-yl]benzoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
6MSN
 
 | |
2YXJ
 
 | | Crystal structure of Bcl-xL in complex with ABT-737 | | Descriptor: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, Apoptosis regulator Bcl-X, CHLORIDE ION, ... | | Authors: | Czabotar, P.E, Lee, E.F, Smith, B.J, Deshayes, K, Zobel, K, Fairlie, W.D, Colman, P.M. | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family
Cell Death Differ., 14, 2007
|
|
1NJR
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase | | Descriptor: | 32.1 kDa protein in ADH3-RCA1 intergenic region, Xylitol | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-02 | | Release date: | 2004-08-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme
Protein Sci., 14, 2005
|
|
6MO4
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50067 complex | | Descriptor: | MAGNESIUM ION, N-[(2R)-1-(hydroxyamino)-3-methyl-3-(methylsulfonyl)-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
