6NWA
 
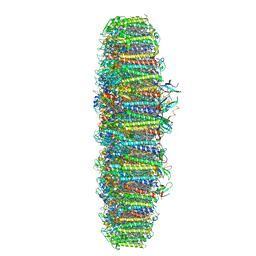 | | The structure of the photosystem I IsiA super-complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Toporik, H, Li, J, Williams, D, Chiu, P.L, Mazor, Y. | | Deposit date: | 2019-02-06 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The structure of the stress-induced photosystem I-IsiA antenna supercomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6VYB
 
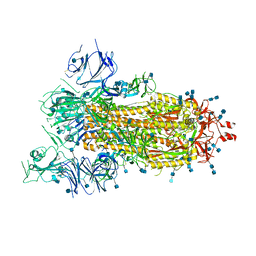 | | SARS-CoV-2 spike ectodomain structure (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Wall, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID), McGuire, A.T, Veesler, D. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein.
Cell, 181, 2020
|
|
8P2G
 
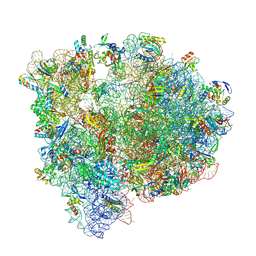 | | Staphylococcus aureus 70S ribosome with elongation factor G locked with fusidic acid cyclopentane with a tRNA in pe/E chimeric state | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 2-[(3~{R},4~{S},5~{S},8~{S},9~{S},10~{S},11~{R},13~{S},14~{S},16~{S})-16-acetyloxy-4,8,10,14-tetramethyl-3,11-bis(oxidanyl)-1,2,3,4,5,6,7,9,11,12,13,15,16,17-tetradecahydrocyclopenta[a]phenanthren-17-yl]-5-cyclopentyl-pentanoic acid, ... | | Authors: | Gonzalez-Lopez, A, Selmer, M. | | Deposit date: | 2023-05-16 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Structures of the Staphylococcus aureus ribosome inhibited by fusidic acid and fusidic acid cyclopentane.
Sci Rep, 14, 2024
|
|
8G6W
 
 | |
8R0B
 
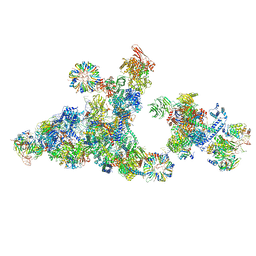 | | Cryo-EM structure of the cross-exon pre-B+ATP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8DLP
 
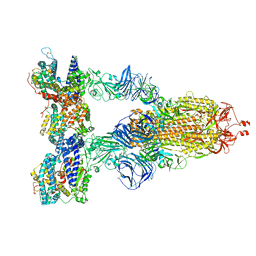 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8OUI
 
 | | Complex of ASCT2 with Suppressyn | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Suppressyn | | Authors: | Khare, S, Kumar, A, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
6J40
 
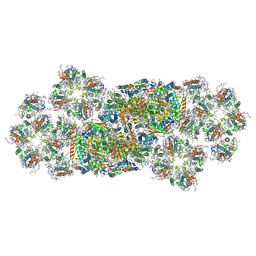 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
8G6Y
 
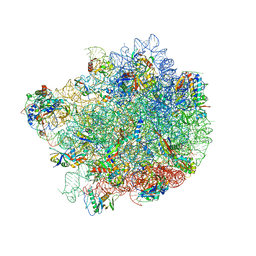 | |
6O3M
 
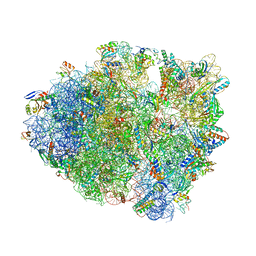 | | Unmodified tRNA(Pro) bound to Thermus thermophilus 70S (cognate) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-02-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
8R08
 
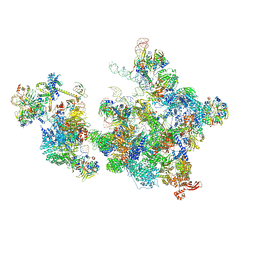 | | Cryo-EM structure of the cross-exon pre-B+AMPPNP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8DUA
 
 | | SIV E660.CR54 SOS-2P Env Trimer with ITS92.02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, Envelope glycoprotein gp41, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of prefusion SIV envelope trimer.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
7U6R
 
 | |
8R9W
 
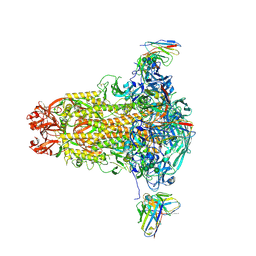 | |
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
6O97
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with propylamycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.75A resolution | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxyc yclohexyl 2-amino-2,4-dideoxy-4-propyl-alpha-D-glucopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Matsushita, T, Sati, G.C, Kondasinghe, N, Pirrone, M.G, Kato, T, Waduge, P, Kumar, H.S, Sanchon, A.C, Dobosz-Bartoszek, M, Shcherbakov, D, Juhas, M, Hobbie, S.N, Schrepfer, T, Chow, C.S, Polikanov, Y.S, Schacht, J, Vasella, A, Bottger, E.C, Crich, D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens.
J. Am. Chem. Soc., 141, 2019
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
8GHU
 
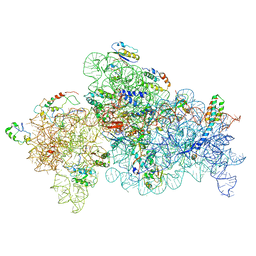 | |
7TUX
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with [(3S)-4-Hydroxy-3-[({2-amino-4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl}methyl)amino]butyl]phosphonic acid | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, ... | | Authors: | Harijan, R.K, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-03 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition and Mechanism of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase.
Acs Chem.Biol., 17, 2022
|
|
8S1P
 
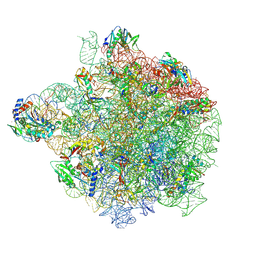 | | YlmH bound to PtRNA-50S | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2024-02-15 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A role for the S4-domain containing protein YlmH in ribosome-associated quality control in Bacillus subtilis.
Nucleic Acids Res., 52, 2024
|
|
8IYG
 
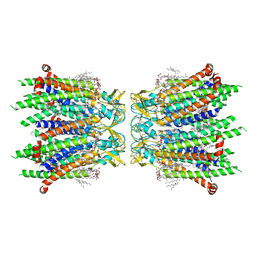 | | Human neuronal gap junction channel connexin 36 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, Gap junction delta-2 protein | | Authors: | Mao, W.X, Chen, S.S. | | Deposit date: | 2023-04-04 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Assembly mechanisms of the neuronal gap junction channel connexin 36 elucidated by Cryo-EM.
Arch.Biochem.Biophys., 754, 2024
|
|
6O65
 
 | |
6JBS
 
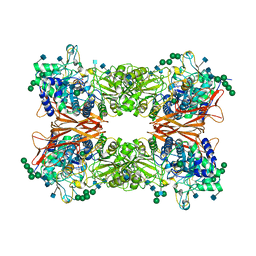 | | Bifunctional xylosidase/glucosidase LXYL | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-01-26 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
