1I13
 
 | | ANALYSIS OF AN INVARIANT ASPARTIC ACID IN HPRTS-ALANINE MUTANT | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, FORMIC ACID, ... | | Authors: | Canyuk, B, Focia, P.J, Eakin, A.E. | | Deposit date: | 2001-01-30 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography.
Biochemistry, 40, 2001
|
|
1I14
 
 | | ANALYSIS OF AN INVARIANT ASPARTIC ACID IN HPRTS-GLUTAMIC ACID MUTANT | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Canyuk, B, Focia, P.J, Eakin, A.E. | | Deposit date: | 2001-01-30 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography.
Biochemistry, 40, 2001
|
|
4QBZ
 
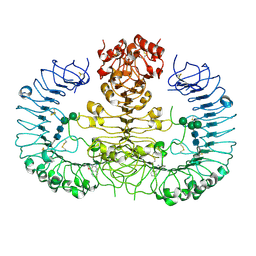 | | Crystal structure of human TLR8 in complex with DS-802 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-butyl[1,3]oxazolo[4,5-c]quinolin-4-amine, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of Activity at Human Toll-like Receptors 7 and 8: Quantitative Structure-Activity Relationship (QSAR) of Diverse Heterocyclic Scaffolds
J.Med.Chem., 57, 2014
|
|
8A2H
 
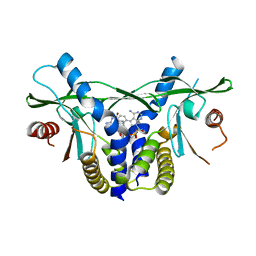 | | human STING in complex with 2',3'-cyclic-GMP-7-deazaphenyl-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanyl-5-phenyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Smola, M, Vavrina, Z, Boura, E, Brynda, J. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2J
 
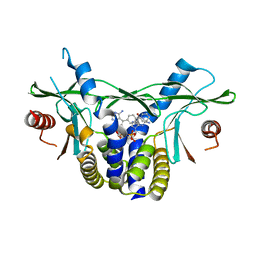 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-biphenylyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-phenylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A1A
 
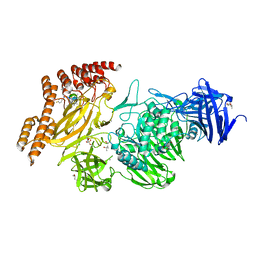 | | Structure of a leucinostatin derivative determined by host lattice display : L1F11V1 construct | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, ... | | Authors: | Mittl, P.R.E. | | Deposit date: | 2022-06-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1TV8
 
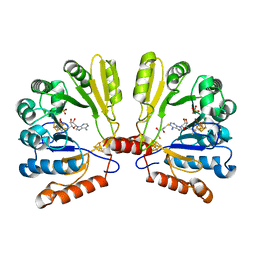 | | Structure of MoaA in complex with S-adenosylmethionine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1LX5
 
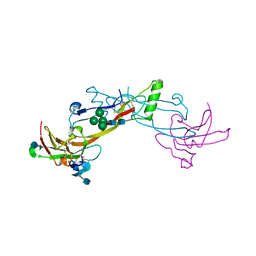 | | Crystal Structure of the BMP7/ActRII Extracellular Domain Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin Type II Receptor, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-04 | | Release date: | 2003-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
2PEH
 
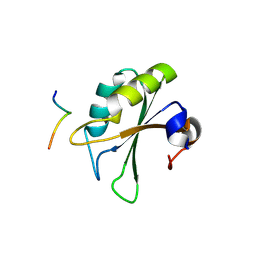 | | Crystal structure of the UHM domain of human SPF45 in complex with SF3b155-ULM5 | | Descriptor: | Splicing factor 3B subunit 1, Splicing factor 45 | | Authors: | Corsini, L, Basquin, J, Hothorn, M, Sattler, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | U2AF-homology motif interactions are required for alternative splicing regulation by SPF45.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6WYE
 
 | | Crystal structure of Neisseria gonorrhoeae serine acetyltransferase (CysE) | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SODIUM ION, Serine acetyltransferase | | Authors: | Hicks, J.L, Oldham, K.E, Summers, E.L, Prentice, E.J. | | Deposit date: | 2020-05-12 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Serine acetyltransferase from Neisseria gonorrhoeae; structural and biochemical basis of inhibition.
Biochem.J., 479, 2022
|
|
1QQ0
 
 | | COBALT SUBSTITUTED CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-10 | | Release date: | 1999-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
5PWD
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 in complex with N09600b | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(methylamino)benzoic acid, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7AD0
 
 | | X-ray structure of Mdm2 with modified p53 peptide | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Modified p53 peptide | | Authors: | Twarda-Clapa, A, Fortuna, P, Grudnik, P, Dubin, G, Berlicki, L, Holak, T.A. | | Deposit date: | 2020-09-13 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Systematic ""foldamerization"" of peptide inhibiting p53-MDM2/X interactions by the incorporation of trans- or cis-2-aminocyclopentanecarboxylic acid residues
Eur.J.Med.Chem., 208, 2020
|
|
5MKR
 
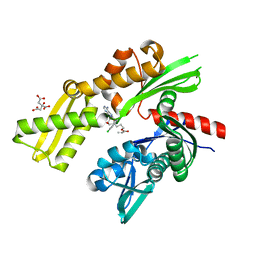 | | HSP72-NBD bound to compound TCI 8 - Tyr15 in up-conformation | | Descriptor: | 3-[(2~{R},3~{S},4~{R},5~{R})-5-[6-azanyl-8-[(4-chlorophenyl)methylamino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propyl prop-2-enoate, CITRATE ANION, Heat shock 70 kDa protein 1A | | Authors: | Pettinger, J, Westwood, I.M, Cronin, N, Le Bihan, Y.-V, Van Montfort, R.L.M. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An Irreversible Inhibitor of HSP72 that Unexpectedly Targets Lysine-56.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5BMV
 
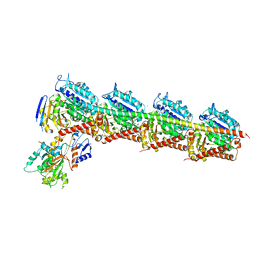 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-Vinblastine COMPLEX | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Chen, Q, Zhang, R. | | Deposit date: | 2015-05-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules.
Mol.Pharmacol., 89, 2016
|
|
3C1Z
 
 | |
6XT7
 
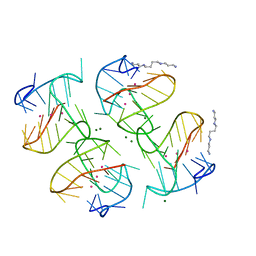 | | Tel25 Hybrid Four-quartet G-quadruplex with K+ | | Descriptor: | DNA (25-MER), MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Yatsunyk, L.A, McCarthy, S.E. | | Deposit date: | 2020-07-17 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The first crystal structures of hybrid and parallel four-tetrad intramolecular G-quadruplexes.
Nucleic Acids Res., 50, 2022
|
|
3GN2
 
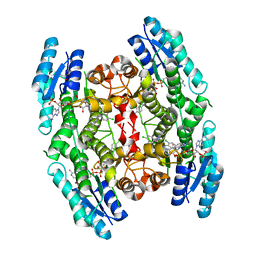 | | Structure of Pteridine Reductase 1 (PTR1) from TRYPANOSOMA BRUCEI in ternary complex with cofactor (NADP+) and inhibitor (DDD00066730) | | Descriptor: | 1-(3,4-dichlorobenzyl)-1H-benzimidazol-2-amine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Brenk, R, Hunter, W.N. | | Deposit date: | 2009-03-16 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment hits discovered by virtual screening.
J.Med.Chem., 52, 2009
|
|
1P0K
 
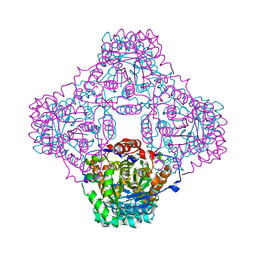 | | IPP:DMAPP isomerase type II apo structure | | Descriptor: | Isopentenyl-diphosphate delta-isomerase, PHOSPHATE ION | | Authors: | Steinbacher, S, Kaiser, J, Gerhardt, S, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Type II Isopentenyl Diphosphate:Dimethylallyl Diphosphate Isomerase from Bacillus subtilis
J.Mol.Biol., 329, 2003
|
|
6UNI
 
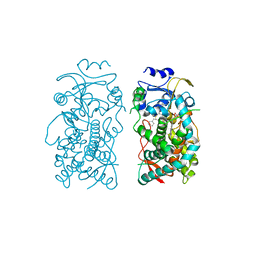 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
1W7V
 
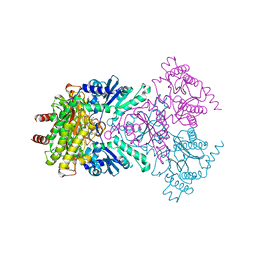 | | ZnMg substituted aminopeptidase P from E. coli | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PEPTIDE VAL-PRO-LEU, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-09-13 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
8CR0
 
 | | Crystal structure of human carbonic anhydrase II in complex with a triazolyl benzoxaborole inhibitor | | Descriptor: | 1-[1,1-bis(oxidanyl)-3~{H}-2,1$l^{4}-benzoxaborol-6-yl]-4-[(4-fluoranylphenoxy)methyl]-1,2,3-triazole, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | 6-Substituted Triazolyl Benzoxaboroles as Selective Carbonic Anhydrase Inhibitors: In Silico Design, Synthesis, and X-ray Crystallography.
J.Med.Chem., 66, 2023
|
|
7QTT
 
 | |
3ZGO
 
 | |
5MKS
 
 | | HSP72-NBD bound to compound TCI 8 - Tyr15 in down-conformation | | Descriptor: | 3-[(2~{R},3~{S},4~{R},5~{R})-5-[6-azanyl-8-[(4-chlorophenyl)methylamino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propyl prop-2-enoate, Heat shock 70 kDa protein 1A | | Authors: | Pettinger, J, Westwood, I.M, Cronin, N, Le Bihan, Y.-V, Van Montfort, R.L.M. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An Irreversible Inhibitor of HSP72 that Unexpectedly Targets Lysine-56.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
