1US5
 
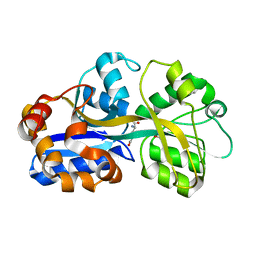 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2Q3T
 
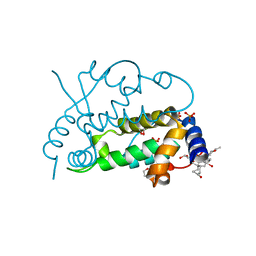 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Protein At3g22680, ... | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
5CG5
 
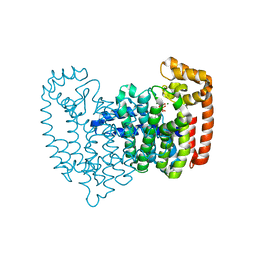 | | Neutron crystal structure of human farnesyl pyrophosphate synthase in complex with risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Yokoyama, T, Mizuguchi, M, Ostermann, A, Kusaka, K, Niimura, N, Schrader, T.E, Tanaka, I. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-14 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.402 Å), X-RAY DIFFRACTION | | Cite: | Protonation State and Hydration of Bisphosphonate Bound to Farnesyl Pyrophosphate Synthase
J.Med.Chem., 58, 2015
|
|
1PY5
 
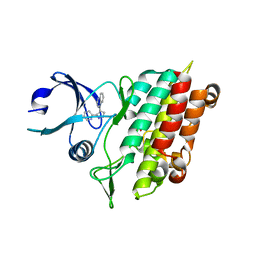 | | Crystal Structure of TGF-beta receptor I kinase with inhibitor | | Descriptor: | 4-(3-PYRIDIN-2-YL-1H-PYRAZOL-4-YL)QUINOLINE, SULFATE ION, TGF-beta receptor type I | | Authors: | Zhang, F, Sawyer, J.S. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
6RRY
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-1,2,3,6-tetrahydro-3-pyridinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)-1,2,3,6-tetrahydropyridin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
4Y8D
 
 | | Crystal structure of Cyclin-G associated kinase (GAK) complexed with selective 12i inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-[3-(morpholin-4-yl)[1,2]thiazolo[4,3-b]pyridin-6-yl]aniline, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Heroven, C, Nowak, R, De Jonghe, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
J.Med.Chem., 58, 2015
|
|
5HIA
 
 | | Human hypoxanthine-guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((S)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Guddat, L.W, Keough, D.T, Rejman, D. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 2017
|
|
4HVD
 
 | | JAK3 kinase domain in complex with 2-Cyclopropyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1,2,2-trimethyl-propyl)-amide | | Descriptor: | 1-phenylurea, 2-cyclopropyl-N-[(2S)-3,3-dimethylbutan-2-yl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 3-Amido Pyrrolopyrazine JAK Kinase Inhibitors: Development of a JAK3 vs JAK1 Selective Inhibitor and Evaluation in Cellular and in Vivo Models.
J.Med.Chem., 56, 2013
|
|
5C8I
 
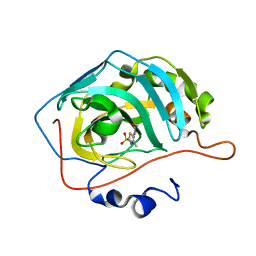 | | Joint X-ray/neutron structure of Human Carbonic Anhydrase II in complex with Methazolamide | | Descriptor: | Carbonic anhydrase 2, N-(3-methyl-5-sulfamoyl-1,3,4-thiadiazol-2(3H)-ylidene)acetamide, ZINC ION | | Authors: | Aggarwal, M, Kovalevsky, A.Y, Fisher, S.Z, McKenna, R. | | Deposit date: | 2015-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.56 Å), X-RAY DIFFRACTION | | Cite: | Neutron structure of human carbonic anhydrase II in complex with methazolamide: mapping the solvent and hydrogen-bonding patterns of an effective clinical drug.
IUCrJ, 3, 2016
|
|
4YD4
 
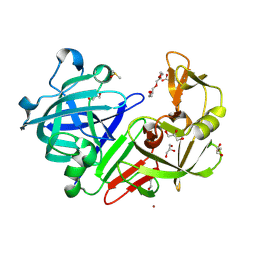 | |
4I9T
 
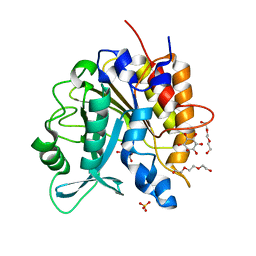 | | Structure of the H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, SULFATE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
2YGW
 
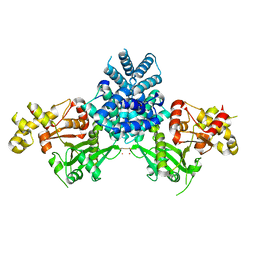 | | Crystal structure of human MCD | | Descriptor: | 1,2-ETHANEDIOL, MALONYL-COA DECARBOXYLASE, MITOCHONDRIAL, ... | | Authors: | Vollmar, M, Puranik, S, Krojer, T, Savitsky, P, Allerston, C, Yue, W.W, Chaikuad, A, von Delft, F, Gileadi, O, Kavanagh, K, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Malonyl-Coenzyme a Decarboxylase Provide Insights Into its Catalytic Mechanism and Disease-Causing Mutations.
Structure, 21, 2013
|
|
4YEK
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6VEB
 
 | | Precorrin-2-bound S128A S. typhimurium siroheme synthase | | Descriptor: | 3,3',3'',3'''-[(7S,8S,12S,13S)-3,8,13,17-tetrakis(carboxymethyl)-8,13-dimethyl-7,8,12,13,20,24-hexahydroporphyrin-2,7,1 2,18-tetrayl]tetrapropanoic acid, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2019-12-31 | | Release date: | 2020-03-04 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site.
Nat Commun, 11, 2020
|
|
1XY9
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
6CEH
 
 | | Design, Synthesis, X-ray and Biological Activities of Selenides Bearing the Benzenesulfonamide Moiety as New Class of Agents for Prevention of Diabetic Cerebrovascular Pathology | | Descriptor: | 4-[(prop-2-en-1-yl)selanyl]benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, di Cesare Mannelli, L, Trallori, E, Ghelardini, C, Carta, F, Supuran, C.T. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Design, Synthesis, and X-ray of Selenides as New Class of Agents for Prevention of Diabetic Cerebrovascular Pathology.
ACS Med Chem Lett, 9, 2018
|
|
6JAR
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R49A in complex with maltose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
8JBY
 
 | | Vascular endothelial protein tyrosine phosphatase in complex with Cpd-2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(hydroxymethyl)-5-(1-methylindol-3-yl)-1,2-oxazole-3-carboxylic acid, ... | | Authors: | Orita, T, Furuzono, T, Doi, S, Adachi, T. | | Deposit date: | 2023-05-10 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Novel VE-PTP Inhibitors Using Orthogonal Biophysical Techniques.
Biochemistry, 62, 2023
|
|
1V5S
 
 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3
To be Published
|
|
6FYR
 
 | | X-RAY STRUCTURE OF CLK3-KD(GP-[275-632], NON-PHOS.)/Cpd-2 AT 1.42A | | Descriptor: | 6-~{tert}-butyl-~{N}-[6-(1~{H}-pyrazol-4-yl)-1~{H}-imidazo[1,2-a]pyridin-2-yl]pyridine-3-carboxamide, Dual specificity protein kinase CLK3 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
5XND
 
 | |
5EDG
 
 | |
6RLL
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ44064146 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)-5-(1,8-diazaspiro[4.5]decan-1-ylmethyl)oxolane-3,4-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Brown, D.G, Robinson, C.M, Pande, V. | | Deposit date: | 2019-05-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A chemical probe for the methyl transferase PRMT5 with a novel
binding mode
To Be Published
|
|
2Z6B
 
 | | Crystal Structure Analysis of (gp27-gp5)3 conjugated with Fe(III) protoporphyrin | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, Baseplate structural protein Gp27, Tail-associated lysozyme | | Authors: | Koshiyama, T, Yokoi, N, Ueno, T, Kanamaru, S, Nagano, S, Shiro, Y, Arisaka, F, Watanabe, Y. | | Deposit date: | 2007-07-28 | | Release date: | 2008-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Molecular design of heteroprotein assemblies providing a bionanocup as a chemical reactor.
Small, 4, 2008
|
|
5XC1
 
 | | Crystal structure of the complex of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 with S-1,2-Propanediol | | Descriptor: | Beta-xylanase, MAGNESIUM ION, S-1,2-PROPANEDIOL, ... | | Authors: | Bansia, H, Mahanta, P, Ramakumar, S. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
