6CO1
 
 | |
6MY0
 
 | |
6MXX
 
 | | Structure of 53BP1 tandem Tudor domains in complex with small molecule UNC2991 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-iodobenzamide, PHOSPHATE ION, ... | | Authors: | Cui, G, Botuyan, M.V, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
4CRI
 
 | | Crystal Structure of 53BP1 tandem tudor domains in complex with methylated K810 Rb peptide | | Descriptor: | RB1 PROTEIN, TUMOR SUPPRESSOR P53-BINDING PROTEIN 1 | | Authors: | Krojer, T, Johansson, C, Gileadi, C, Fedorov, O, Carr, S, La Thangue, N.B, Vollmar, M, Crawley, L, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-02-26 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lysine Methylation-Dependent Binding of 53BP1 to the Prb Tumor Suppressor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CO2
 
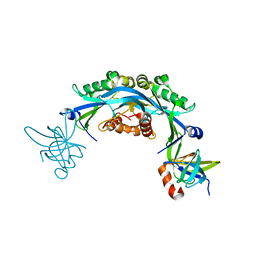 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | Descriptor: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | Authors: | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | Deposit date: | 2018-03-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1KZY
 
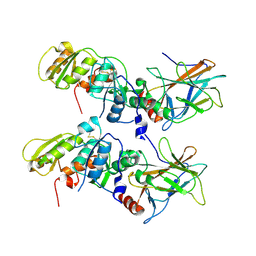 | | Crystal Structure of the 53bp1 BRCT Region Complexed to Tumor Suppressor P53 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, TUMOR SUPPRESSOR P53-BINDING PROTEIN 1, ZINC ION | | Authors: | Joo, W.S, Jeffrey, P.D, Cantor, S.B, Finnin, M.S, Livingston, D.M, Pavletich, N.P. | | Deposit date: | 2002-02-08 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure.
Genes Dev., 16, 2002
|
|
6MXZ
 
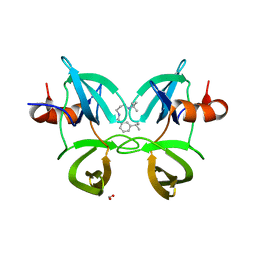 | | Structure of 53BP1 Tudor domains in complex with small molecule UNC3474 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-(propan-2-yl)benzamide, TP53-binding protein 1 | | Authors: | Cui, G, Botuyan, M.V, Schuller, D.J, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
5J26
 
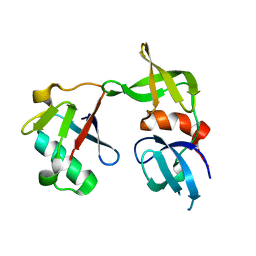 | | Crystal structure of a 53BP1 Tudor domain in complex with a ubiquitin variant | | Descriptor: | Tumor suppressor p53-binding protein 1, Ubiquitin Variant i53 | | Authors: | Wan, L, Canny, M, Juang, Y.C, Durocher, D, Sicheri, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5047 Å) | | Cite: | A genetically encoded inhibitor of 53BP1 to stimulate homology-based gene editing
To Be Published
|
|
1GZH
 
 | |
1XNI
 
 | | Tandem Tudor Domain of 53BP1 | | Descriptor: | Tumor suppressor p53-binding protein 1 | | Authors: | Huyen, Y, Zgheib, O, DiTullio Jr, R.A, Gorgoulis, V.G, Zacharatos, P, Petty, T.J, Sheston, E.A, Mellert, H.S, Stavridi, E.S, Halazonetis, T.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks
Nature, 432, 2004
|
|
6RML
 
 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a 53BP1 phosphopeptide | | Descriptor: | 53BP1, DNA topoisomerase 2-binding protein 1 | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint.
Elife, 8, 2019
|
|
5ECG
 
 | | Crystal structure of the BRCT domains of 53BP1 in complex with p53 and H2AX-pSer139 (gammaH2AX) | | Descriptor: | Cellular tumor antigen p53, SEP-GLN-GLU-TYR, Tumor suppressor p53-binding protein 1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATM Localization and Heterochromatin Repair Depend on Direct Interaction of the 53BP1-BRCT2 Domain with gamma H2AX.
Cell Rep, 13, 2015
|
|
7LIO
 
 | |
7YQK
 
 | | cryo-EM structure of gammaH2AXK15ub-H4K20me2 nucleosome bound to 53BP1 | | Descriptor: | DNA (145-MER), Histone H2AX, Histone H2B, ... | | Authors: | Ai, H.S, GuoChao, C, Qingyue, G, Ze-Bin, T, Zhiheng, D, Xin, L, Fan, Y, Ziyu, X, Jia-Bin, L, Changlin, T, Liu, L. | | Deposit date: | 2022-08-07 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Chemical Synthesis of Post-Translationally Modified H2AX Reveals Redundancy in Interplay between Histone Phosphorylation, Ubiquitination, and Methylation on the Binding of 53BP1 with Nucleosomes.
J.Am.Chem.Soc., 144, 2022
|
|
6RMM
 
 | |
8U4U
 
 | |
5KGF
 
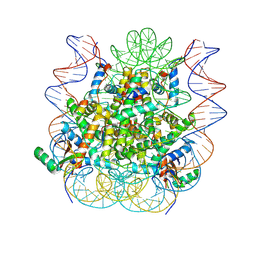 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
2LVM
 
 | |
2MWO
 
 | |
2MWP
 
 | |
