3LE2
 
 | | Structure of Arabidopsis AtSerpin1. Native Stressed Conformation | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Harrop, S.J, Joss, T.V, Cumi, P.M.G, Roberts, T.H. | | Deposit date: | 2010-01-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arabidopsis AtSerpin1, crystal structure and in vivo interaction with its target protease RESPONSIVE TO DESICCATION-21 (RD21).
J.Biol.Chem., 285, 2010
|
|
3LW2
 
 | | Mouse Plasminogen Activator Inhibitor-1 (PAI-1) | | Descriptor: | Plasminogen activator inhibitor 1 | | Authors: | Dewilde, M, Van De Craen, B, Compernolle, G, Madsen, J.B, Strelkov, S.V, Gils, A, Declerck, P.J. | | Deposit date: | 2010-02-23 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Subtle structural differences between human and mouse PAI-1 reveal the basis for biochemical differences.
J.Struct.Biol., 171, 2010
|
|
6ZRV
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) in Complex with an Inhibitory Nanobody (VHH-s-a93, Nb93) | | Descriptor: | Plasminogen activator inhibitor 1, VHH-s-a93 (Nb93) | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity.
Int J Mol Sci, 21, 2020
|
|
4EB1
 
 | | Hyperstable in-frame insertion variant of antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III | | Authors: | Martinez-Martinez, I, Johnson, D.J.D, Yamasaki, M, Corral, J, Huntington, J.A. | | Deposit date: | 2012-03-23 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Type II antithrombin deficiency caused by a large in-frame insertion: structural, functional and pathological relevance.
J.Thromb.Haemost., 10, 2012
|
|
7API
 
 | | THE S VARIANT OF HUMAN ALPHA1-ANTITRYPSIN, STRUCTURE AND IMPLICATIONS FOR FUNCTION AND METABOLISM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTITRYPSIN, ... | | Authors: | Loebermann, H, Tokuoka, R, Deisenhofer, J, Huber, R. | | Deposit date: | 1988-09-08 | | Release date: | 1990-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism.
Protein Eng., 2, 1989
|
|
7AQF
 
 | |
7AQG
 
 | | Crystal Structure of Small Molecule Inhibitor TM5484 Bound to Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) | | Descriptor: | 5-Chloro-2-[[2-[3-(furan-3-yl)anilino]-2-oxoacetyl]amino]benzoic acid, Plasminogen activator inhibitor 1, VHH-2g-42 (Nb42), ... | | Authors: | Sillen, M, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Insight into the Two-Step Mechanism of PAI-1 Inhibition by Small Molecule TM5484.
Int J Mol Sci, 22, 2021
|
|
7AEL
 
 | | alpha 1-antitrypsin (C232S) complexed with GSK716 | | Descriptor: | Alpha-1-antitrypsin, SULFATE ION, ~{N}-[(1~{S},2~{R})-1-(3-fluoranyl-2-methyl-phenyl)-1-oxidanyl-pentan-2-yl]-2-oxidanylidene-1,3-dihydroindole-4-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of a small molecule that corrects misfolding and increases secretion of Z alpha 1 -antitrypsin.
Embo Mol Med, 13, 2021
|
|
7AHP
 
 | | Crystal structure of Ixodes ricinus serpin - Iripin-3 | | Descriptor: | Putative salivary serpin, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Kascakova, B, Kuta Smatanova, I, Prudnikova, T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Iripin-3, a New Salivary Protein Isolated From Ixodes ricinus Ticks, Displays Immunomodulatory and Anti-Hemostatic Properties In Vitro
Front Immunol, 12, 2021
|
|
4G8R
 
 | | Crystal Structure of a novel small molecule inactivator bound to plasminogen activator inhibitor-1 | | Descriptor: | (2S)-3-({[3-(trifluoromethyl)phenoxy]carbonyl}amino)propane-1,2-diyl bis(3,4,5-trihydroxybenzoate), Plasminogen activator inhibitor-1, SULFATE ION | | Authors: | Stuckey, J.A, Lawrence, D.A, Li, S.-H. | | Deposit date: | 2012-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mechanistic characterization and crystal structure of a small molecule inactivator bound to plasminogen activator inhibitor-1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3NDA
 
 | | Crystal structure of serpin from tick Ixodes ricinus | | Descriptor: | PENTAETHYLENE GLYCOL, Serpin-2 | | Authors: | Rezacova, P, Kovarova, Z, Chmelar, J, Mares, M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation.
Blood, 117, 2011
|
|
3NE4
 
 | |
3NDF
 
 | |
3NDD
 
 | |
3Q02
 
 | |
3PZF
 
 | | 1.75A resolution structure of Serpin-2 from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, An, C, Michel, K. | | Deposit date: | 2010-12-14 | | Release date: | 2011-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of native Anopheles gambiae serpin-2, a negative regulator of melanization in mosquitoes.
Proteins, 79, 2011
|
|
3OZQ
 
 | |
3Q03
 
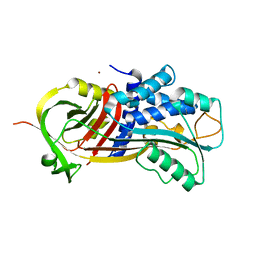 | |
3R4L
 
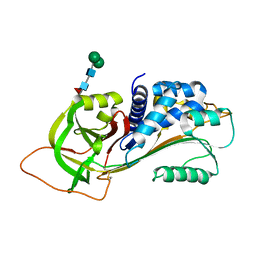 | | Human very long half life Plasminogen Activator Inhibitor type-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasminogen activator inhibitor 1, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Zheng, H, Han, Q, Skrzypczak-Jankun, E, Jankun, J. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Remarkable extension of PAI-1 half-life surprisingly brings no changes to its structure.
Int.J.Mol.Med., 29, 2012
|
|
1YXA
 
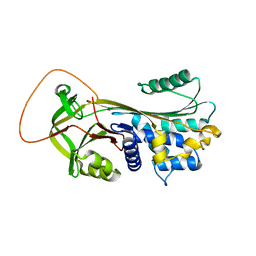 | | Serpina3n, a murine orthologue of human antichymotrypsin | | Descriptor: | serine (or cysteine) proteinase inhibitor, clade A, member 3N | | Authors: | Horvath, A.J, Irving, J.A, Law, R.H, Rossjohn, J, Bottomley, S.P, Quinsey, N.S, Pike, R.N, Coughlin, P.B, Whisstock, J.C. | | Deposit date: | 2005-02-20 | | Release date: | 2005-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins.
J.Biol.Chem., 280, 2005
|
|
1LJ5
 
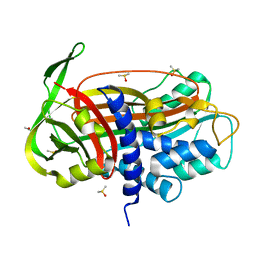 | |
1M93
 
 | |
2ARR
 
 | | Human plasminogen activator inhibitor-2.[loop (66-98) deletion mutant] complexed with peptide n-acetyl-teaaagmggvmtgr-oh | | Descriptor: | 14-mer from Plasminogen activator inhibitor-2, Plasminogen activator inhibitor-2 | | Authors: | Di Giusto, D.A, Sutherland, A.P, Jankova, L, Harrop, S.J, Curmi, P.M, King, G.C. | | Deposit date: | 2005-08-21 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Plasminogen activator inhibitor-2 is highly tolerant to P8 residue substitution--implications for serpin mechanistic model and prediction of nsSNP activities
J.Mol.Biol., 353, 2005
|
|
1LK6
 
 | | Structure of dimeric antithrombin complexed with a P14-P9 reactive loop peptide and an exogenous tripeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Carrell, R.W, Stein, P.E. | | Deposit date: | 2002-04-24 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serpin Polymerization Is Prevented by a Hydrogen Bond Network That Is Centered on His-334 and Stabilized by Glycerol
J.Biol.Chem., 278, 2003
|
|
2ACH
 
 | | CRYSTAL STRUCTURE OF CLEAVED HUMAN ALPHA1-ANTICHYMOTRYPSIN AT 2.7 ANGSTROMS RESOLUTION AND ITS COMPARISON WITH OTHER SERPINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTICHYMOTRYPSIN, PHOSPHATE ION, ... | | Authors: | Baumann, U, Huber, R, Bode, W, Grosse, D, Lesjak, M, Laurell, C.B. | | Deposit date: | 1993-04-26 | | Release date: | 1993-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cleaved human alpha 1-antichymotrypsin at 2.7 A resolution and its comparison with other serpins.
J.Mol.Biol., 218, 1991
|
|
