4LWW
 
 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-(4-(phenylsulfonyl)benzyl)-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | Deposit date: | 2013-07-28 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4LV9
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 7-chloro-3-methyl-2H-1,2,4-benzothiadiazine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LVF
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-2-phenyl-N-(pyridin-4-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ZY4
 
 | |
7VIB
 
 | | Crystal structure of human ACE2 and GX/P2V RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein, ZINC ION | | Authors: | Guo, Y, Cao, W, Jia, N, Wang, W, Yuan, S, Wang, Y. | | Deposit date: | 2021-09-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human ACE2 and GX/P2V RBD
To Be Published
|
|
5BMS
 
 | |
4ZY6
 
 | |
4ZY5
 
 | |
3NBI
 
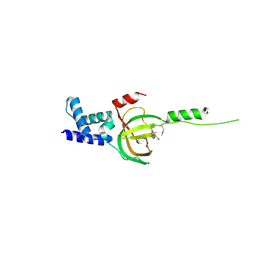 | | Crystal structure of human RMI1 N-terminus | | Descriptor: | RecQ-mediated genome instability protein 1 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
4B5X
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase (HpaI), mutant D42A | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4KFN
 
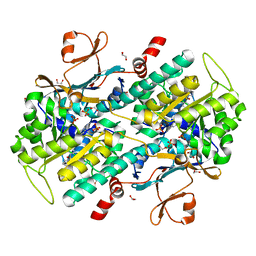 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4B5T
 
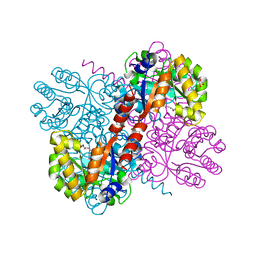 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with ketobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5V
 
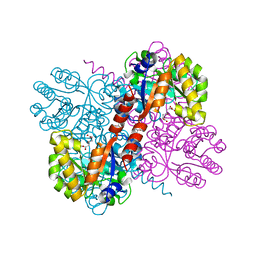 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with 4-hydroxyl-2-ketoheptane-1,7-dioate | | Descriptor: | (4R)-4-oxidanyl-2-oxidanylidene-heptanedioic acid, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, GLYCEROL, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5U
 
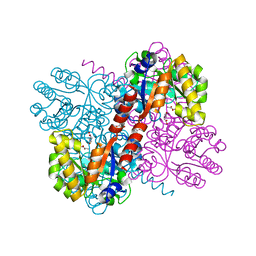 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate and succinic semialdehyde | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, 4-oxobutanoic acid, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5W
 
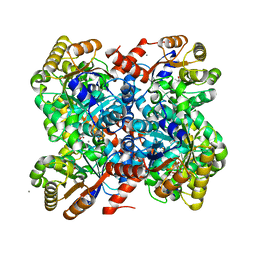 | | Crystal structures of divalent metal dependent pyruvate aldolase R70A mutant, HpaI, in complex with pyruvate | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
6URC
 
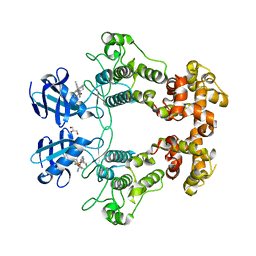 | | Crystal structure of IRE1a in complex with compound 18 | | Descriptor: | 2-chloro-N-(6-methyl-5-{[3-(2-{[(3S)-piperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}naphthalen-1-yl)benzene-1-sulfonamide, GLYCEROL, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.H, Wang, W. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of IRE1 alpha through its kinase domain attenuates multiple myeloma.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4B5S
 
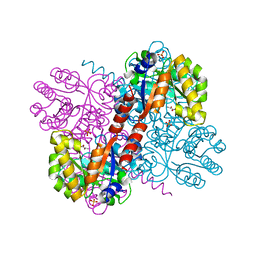 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, D-Glyceraldehyde, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4Q5W
 
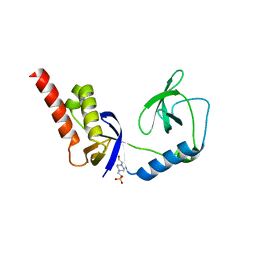 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4KFO
 
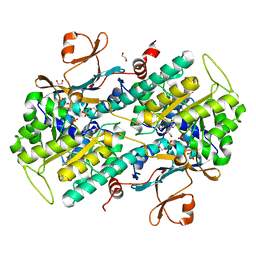 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Tosteb, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
6W3E
 
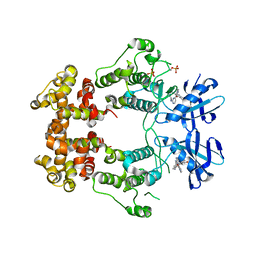 | | Structure of phosphorylated IRE1 in complex with G-0701 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, methyl ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]carbamate | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.737 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
6W39
 
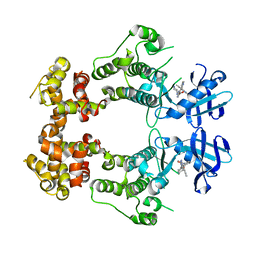 | | Structure of unphosphorylated IRE1 in complex with G-1749 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, ethyl ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]carbamate | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
6W3B
 
 | | Structure of apo unphosphorylated IRE1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H, Mortara, K, Ferri, E, Rudolph, J, Wang, W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
4Q5Y
 
 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4DSO
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, BENZAMIDINE, GLYCEROL, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSU
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | BENZIMIDAZOLE, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
