1NYM
 
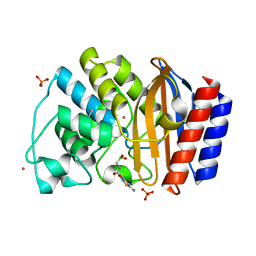 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (CXB) | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1PSJ
 
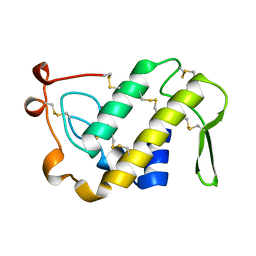 | | ACIDIC PHOSPHOLIPASE A2 FROM AGKISTRODON HALYS PALLAS | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Wang, X.Q, Lin, Z.J. | | Deposit date: | 1995-05-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an acidic phospholipase A2 from the venom of Agkistrodon halys pallas at 2.0 A resolution.
J.Mol.Biol., 255, 1996
|
|
1NYY
 
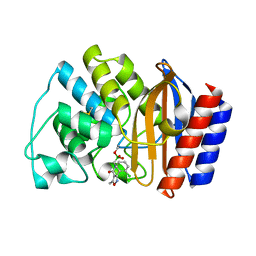 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (105) | | Descriptor: | Beta-lactamase TEM, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-14 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1NY0
 
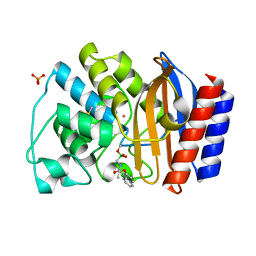 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (NBF) | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-11 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition and Resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1JWV
 
 | |
1JWZ
 
 | |
1AWP
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-10-03 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The reduction potential of cytochrome b5 is modulated by its exposed heme edge.
Biochemistry, 37, 1998
|
|
8K69
 
 | | Cryo-EM structure of Oryza sativa HKT2;2/1 at 2.3 angstrom | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Wang, X, Shen, X, Qu, Y, Wang, C, Shen, H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters.
Nat.Plants, 10, 2024
|
|
8K66
 
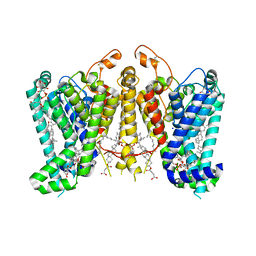 | | Cryo-EM structure of Oryza sativa HKT2;1 at 2.5 angstrom | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Wang, X, Shen, X, Qu, Y, Wang, C, Shen, H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters.
Nat.Plants, 10, 2024
|
|
1IB2
 
 | |
1BML
 
 | |
8JD1
 
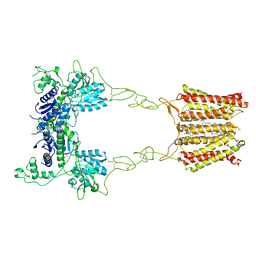 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in Rco state | | Descriptor: | CHOLESTEROL, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCW
 
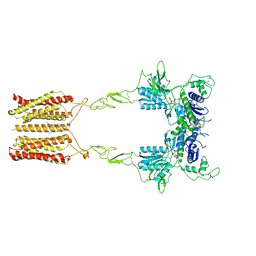 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD2
 
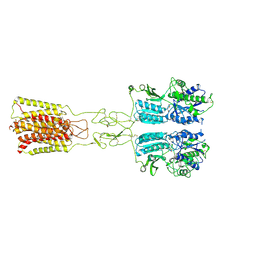 | | Cryo-EM structure of G protein-free mGlu2-mGlu3 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCV
 
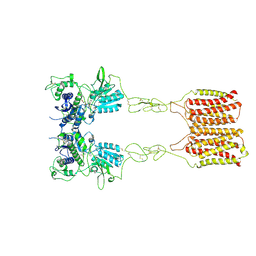 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCZ
 
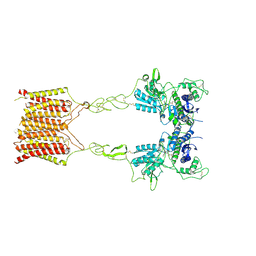 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode III) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD6
 
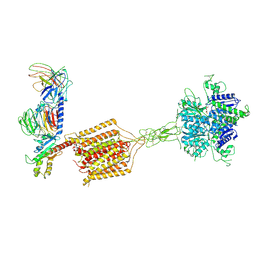 | | Cryo-EM structure of Gi1-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCY
 
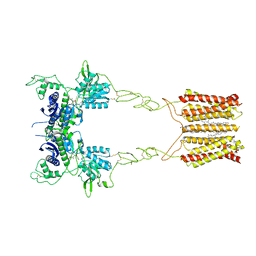 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD0
 
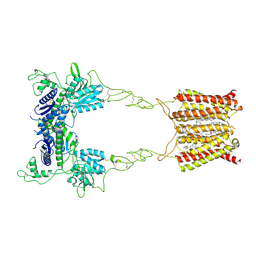 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of NAM563 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCU
 
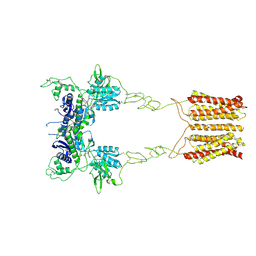 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCX
 
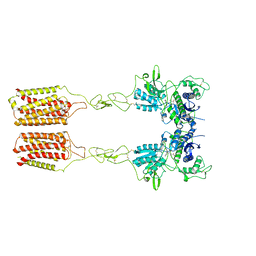 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD5
 
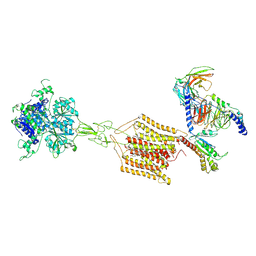 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu4 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-N-(4-methylpyrimidin-2-yl)-4-(1H-pyrazol-4-yl)-1,3-thiazol-2-amine, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD3
 
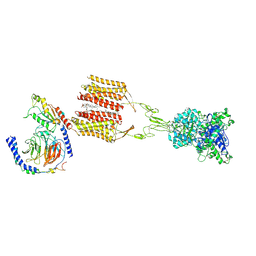 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu3 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, CHOLESTEROL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD4
 
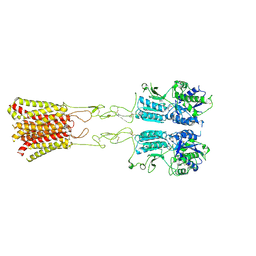 | | Cryo-EM structure of G protein-free mGlu2-mGlu4 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
1DL0
 
 | |
