4OUC
 
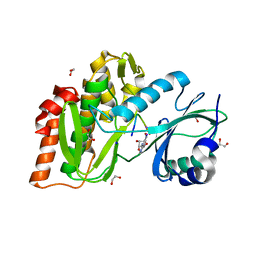 | | Structure of human haspin in complex with histone H3 substrate | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Histone H3.2, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-02-15 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of the chromatin phosphoproteome by the haspin protein kinase.
Mol Cell Proteomics, 13, 2014
|
|
3CXW
 
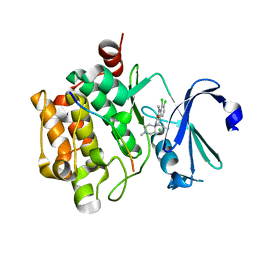 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand I | | Descriptor: | (4R)-7,8-dichloro-1',9-dimethyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, CHLORIDE ION, Pimtide peptide, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, Roos, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
3CY2
 
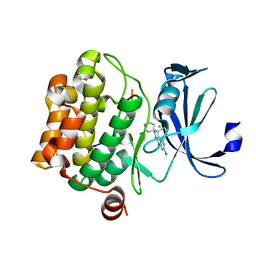 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand II | | Descriptor: | (4R)-7-chloro-9-methyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
3CYN
 
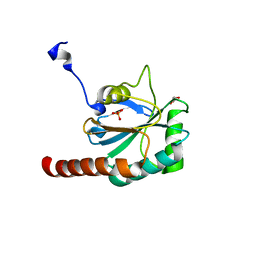 | | The structure of human GPX8 | | Descriptor: | GLYCEROL, Probable glutathione peroxidase 8, SULFATE ION | | Authors: | Kavanagh, K.L, Johansson, C, Yue, W.W, Kochan, G, Pike, A.C.W, Murray, J, Roos, A.K, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GPX8
To be Published
|
|
4OGI
 
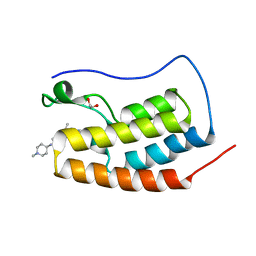 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor BI-2536 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Jose, B, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Dual kinase-bromodomain inhibitors for rationally designed polypharmacology.
Nat.Chem.Biol., 10, 2014
|
|
9FQ2
 
 | | Poliovirus 3C protease in H32 spacegroup | | Descriptor: | Protease 3C | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Poliovirus 3C protease in H32 spacegroup
To Be Published
|
|
9FWC
 
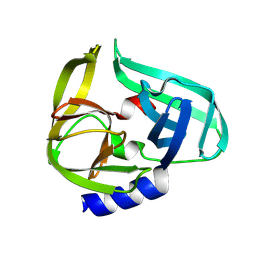 | | Coxsackievirus B3 3C protease in C121 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-28 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Coxsackievirus B3 3C protease in C121 spacegroup
To Be Published
|
|
9FS7
 
 | | Coxsackievirus A16 3C protease in C2 2 21 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R, MacLean, M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Coxsackievirus A16 3C protease in C2 2 21 spacegroup
To Be Published
|
|
9FSB
 
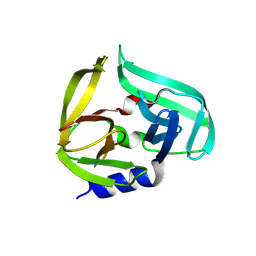 | | Coxsackievirus B3 3C protease in P121 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Coxsackievirus B3 3C protease in P121 spacegroup
To Be Published
|
|
4OGJ
 
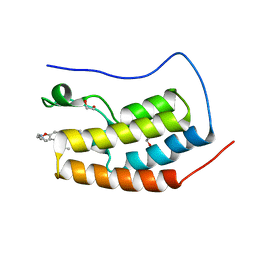 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor TG-101348 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide | | Authors: | Filippakopoulos, P, Picaud, S, Jose, B, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dual kinase-bromodomain inhibitors for rationally designed polypharmacology.
Nat.Chem.Biol., 10, 2014
|
|
8CO8
 
 | | Structure of West Nile Virus NS2B-NS3 protease | | Descriptor: | Serine protease subunit NS2B,Serine protease/Helicase NS3 | | Authors: | Fairhead, M, Godoy, A.S, Koekemoer, L, Balcomb, B.H, Lithgo, R.M, Aschenbrenner, J.C, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-27 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of West Nile Virus NS2B-NS3 protease - to be published
To Be Published
|
|
7PSQ
 
 | | Crystal structure of S100A4 labeled with NU074381b. | | Descriptor: | (2~{R},4~{R})-1-ethanoyl-~{N}-naphthalen-1-yl-4-phenyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Krojer, T, Arruda Bezerra, G, Koekemoer, L, Bountra, C, Von Delft, F, Brennan, P, Fedorov, O. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of S100A4 labeled with NU074381b.
To Be Published
|
|
8CNX
 
 | | Structure of Enterovirus D68 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of EV D68 3C protease
To Be Published
|
|
8CNY
 
 | | Structure of Enterovirus A71 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of EV D68 3C protease - to be published
To Be Published
|
|
2BWJ
 
 | | Structure of adenylate kinase 5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE 5, CHLORIDE ION, ... | | Authors: | Bunkoczi, G, Filippakopoulos, P, Fedorov, O, Jansson, A, Longman, E, Ugochukwu, E, Knapp, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Adenylate Kinase 5
To be Published
|
|
5KCH
 
 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy into weak electron density | | Descriptor: | 4-methoxy-N-[(pyridin-2-yl)methyl]aniline, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
7B9B
 
 | | Crystal structure of human 5' exonuclease Appollo APO form | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-12-14 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
1KS9
 
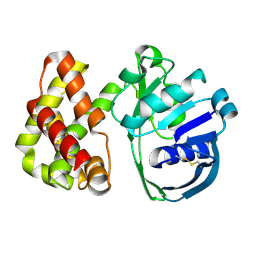 | | Ketopantoate Reductase from Escherichia coli | | Descriptor: | 2-DEHYDROPANTOATE 2-REDUCTASE | | Authors: | Matak-Vinkovic, D, Vinkovic, M, Saldanha, S.A, Ashurst, J.A, von Delft, F, Inoue, T, Miguel, R.N, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2002-01-11 | | Release date: | 2002-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism.
Biochemistry, 40, 2001
|
|
3UW9
 
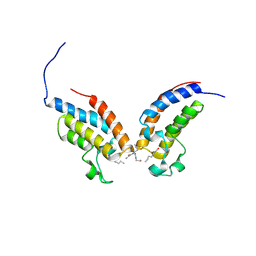 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H4K8acK12ac) | | Descriptor: | Bromodomain-containing protein 4, histone 4 peptide (H4K8acK12ac) | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3D7C
 
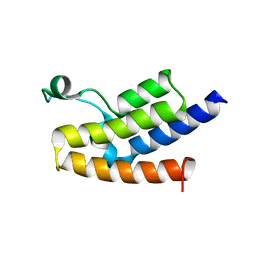 | | Crystal structure of the bromodomain of human GCN5, the general control of amino-acid synthesis protein 5-like 2 | | Descriptor: | General control of amino acid synthesis protein 5-like 2 | | Authors: | Filippakopoulos, P, Eswaran, J, Picaud, S, Fedorov, O, Murray, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-21 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
5KCO
 
 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy | | Descriptor: | DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
3UVX
 
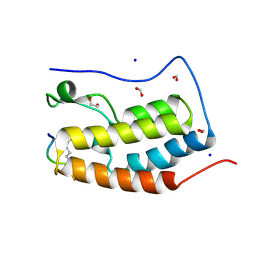 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H4K12acK16ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-30 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3CEK
 
 | | Crystal structure of human dual specificity protein kinase (TTK) | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Filippakopoulos, P, Soundararajan, M, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, Roos, A, Pilka, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
3UV4
 
 | | Crystal Structure of the second bromodomain of human Transcription initiation factor TFIID subunit 1 (TAF1) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
5LAR
 
 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
