8HKN
 
 | |
8HLQ
 
 | |
8HLJ
 
 | |
8HKO
 
 | |
8HKS
 
 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib(BGB-290) | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib
To Be Published
|
|
8HLR
 
 | |
2IS9
 
 | | Structure of yeast DCN-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Defective in cullin neddylation protein 1, ... | | Authors: | Yang, X, Zhou, J, Sun, L, Wei, Z, Gao, J, Gong, W, Xu, R.M, Rao, Z, Liu, Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for the function of DCN-1 in protein Neddylation.
J.Biol.Chem., 282, 2007
|
|
3WFV
 
 | | HIV-1 CRF07 gp41 | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Du, J, Xue, H, Ma, J, Liu, F, Zhou, J, Shao, Y, Qiao, W, Liu, X. | | Deposit date: | 2013-07-24 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of HIV CRF07 B'/C gp41 reveals a hyper-mutant site in the middle of HR2 heptad repeat
Virology, 446, 2013
|
|
3V0A
 
 | | 2.7 angstrom crystal structure of BoNT/Ai in complex with NTNHA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BoNT/A, CALCIUM ION, ... | | Authors: | Gu, S, Rumpel, S, Zhou, J, Strotmeier, J, Bigalke, H, Perry, K, Shoemaker, C.B, Rummel, A, Jin, R. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Botulinum neurotoxin is shielded by NTNHA in an interlocked complex.
Science, 335, 2012
|
|
2HZ6
 
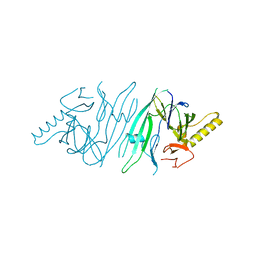 | | The crystal structure of human IRE1-alpha luminal domain | | Descriptor: | Endoplasmic reticulum to nucleus signalling 1 isoform 1 variant | | Authors: | Kaufman, R.J, Xu, Z, Zhou, J. | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3V0B
 
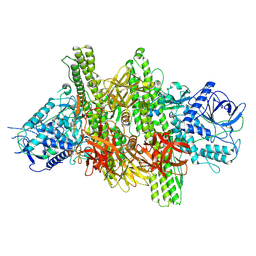 | | 3.9 angstrom crystal structure of BoNT/Ai in complex with NTNHA | | Descriptor: | BoNT/A, CALCIUM ION, NTNH, ... | | Authors: | Gu, S, Rumpel, S, Zhou, J, Strotmeier, J, Bigalke, H, Perry, K, Shoemaker, C.B, Rummel, A, Jin, R. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-14 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Botulinum neurotoxin is shielded by NTNHA in an interlocked complex.
Science, 335, 2012
|
|
3V0C
 
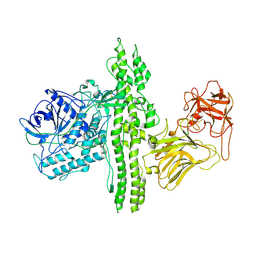 | | 4.3 angstrom crystal structure of an inactive BoNT/A (E224Q/R363A/Y366F) | | Descriptor: | BoNT/A, ZINC ION | | Authors: | Gu, S, Rumpel, S, Zhou, J, Strotmeier, J, Bigalke, H, Perry, K, Shoemaker, C.B, Rummel, A, Jin, R. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Botulinum neurotoxin is shielded by NTNHA in an interlocked complex.
Science, 335, 2012
|
|
5C54
 
 | |
5C55
 
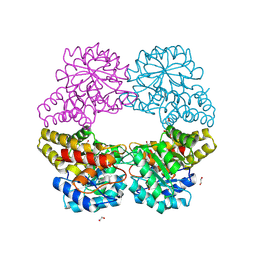 | |
7RST
 
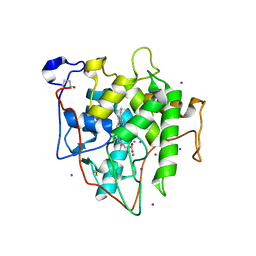 | | The Crystal Structure of Recombinant Chloroperoxidase Expressed in Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chloroperoxidase, ... | | Authors: | Tang, X, Venkadesh, S, Zhou, J, Rosen, B, Wang, X. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The Crystal Structure of Recombinant Chloroperoxidase Expressed in Aspergillus niger
To Be Published
|
|
4QY0
 
 | | Structure of H10 from human-infecting H10N8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY2
 
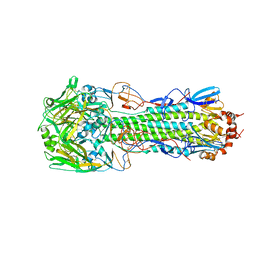 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
3V64
 
 | | Crystal Structure of agrin and LRP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor-related protein 4, ... | | Authors: | Zong, Y, Zhang, B, Gu, S, Lee, K, Zhou, J, Yao, G, Figueiedo, D, Perry, K, Mei, L, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
5GCN
 
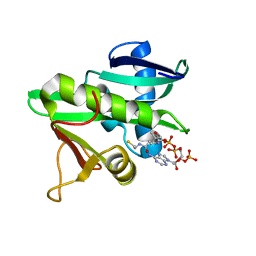 | | CATALYTIC DOMAIN OF TETRAHYMENA GCN5 HISTONE ACETYLTRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | COENZYME A, HISTONE ACETYLTRANSFERASE GCN5 | | Authors: | Lin, Y, Fletcher, C.M, Zhou, J, Allis, C.D, Wagner, G. | | Deposit date: | 1999-03-24 | | Release date: | 1999-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A
Nature, 400, 1999
|
|
5H2T
 
 | |
5H5X
 
 | |
5HSI
 
 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | Descriptor: | MAGNESIUM ION, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
4P5A
 
 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
4MYD
 
 | | 1.37 Angstrom Crystal Structure of E. Coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) in complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-27 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
4MXD
 
 | | 1.45 angstronm crystal structure of E.coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) | | Descriptor: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-26 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
