7WMI
 
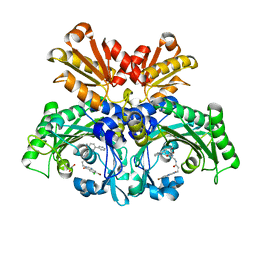 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(1R)-2-[3-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)propanoylamino]-1-(3-phenylphenyl)ethyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Cai, Z, Chen, B, Yu, Y, Zhou, H. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Proof-of-Concept of Triple-Site Inhibitors against Aminoacyl-tRNA Synthetases.
J.Med.Chem., 65, 2022
|
|
7WMF
 
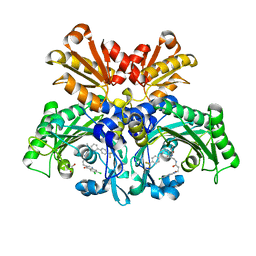 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(1R)-2-[3-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]propanoylamino]-1-[3-[4-(trifluoromethyl)phenyl]phenyl]ethyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Cai, Z, Chen, B, Yu, Y, Zhou, H. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Proof-of-Concept of Triple-Site Inhibitors against Aminoacyl-tRNA Synthetases.
J.Med.Chem., 65, 2022
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8I2J
 
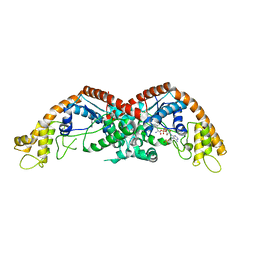 | |
7CBB
 
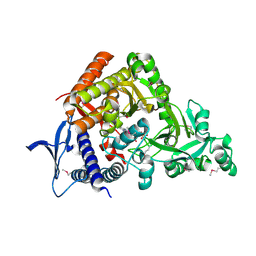 | |
6JIO
 
 | | Human LXR-beta in complex with a ligand | | Descriptor: | Oxysterols receptor LXR-beta, tert-butyl 7-amino-3,4-dihydroisoquinoline-2(1H)-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-02-22 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identify liver X receptor beta modulator building blocks by developing a fluorescence polarization-based competition assay.
Eur.J.Med.Chem., 178, 2019
|
|
6K9H
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-phenyl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
6K9G
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{R},3~{R})-2'-[3-[4-(hydroxymethyl)-3-methylsulfonyl-phenyl]phenyl]-2-oxidanylidene-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
6IDE
 
 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
6K9M
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-propan-2-yl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of novel liver X receptor inverse agonists as lipogenesis inhibitors.
Eur.J.Med.Chem., 206, 2020
|
|
6KJU
 
 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
6A9W
 
 | | Structure of the bifunctional DNA primase-polymerase from phage NrS-1 | | Descriptor: | Primase | | Authors: | Guo, H.J, Li, M.J, Wang, T.L, Wu, H, Zhou, H, Xu, C.Y, Liu, X.P, Yu, F, He, J.H. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and biochemical studies of the bifunctional DNA primase-polymerase from phage NrS-1.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
7TSQ
 
 | | Structure of Enterobacter cloacae Cap2 bound to CdnD02 C-terminus, AMP state | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cap2, Cyclic AMP-AMP-GMP synthase, ... | | Authors: | Ye, Q, Gu, Y, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7TQD
 
 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7TSX
 
 | | Structure of Enterobacter cloacae Cap2 bound to CdnD02 C-terminus, Apo state | | Descriptor: | Cap2, Cyclic AMP-AMP-GMP synthase | | Authors: | Ye, Q, Gu, Y, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7TO3
 
 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7VRB
 
 | | Structure of the Human BRG1/SS18 complex | | Descriptor: | SMARCA4 protein,Protein SSXT | | Authors: | Cheng, Y, Chen, F, Zhou, H, Long, J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Phase transition and remodeling complex assembly are important for SS18-SSX oncogenic activity in synovial sarcomas.
Nat Commun, 13, 2022
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
5ZYQ
 
 | | The Structure of Human PAF1/CTR9 complex | | Descriptor: | RNA polymerase-associated protein CTR9 homolog,RNA polymerase II-associated factor 1 homolog | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
6KNK
 
 | | Crystal structure of SbnH in complex with citryl-diaminoethane | | Descriptor: | (2S)-2-{2-[(2-AMINOETHYL)AMINO]-2-OXOETHYL}-2-HYDROXYBUTANEDIOIC ACID, (2~{S})-2-[2-[2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]ethylamino]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, PHOSPHATE ION, ... | | Authors: | Tang, J, Ju, Y, Zhou, H. | | Deposit date: | 2019-08-05 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
5ZYP
 
 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
6KNH
 
 | | Crystal structure of SbnH in complex with citrate, a PLP-dependent decarboxylase in Staphyloferrin B biothesynthesis | | Descriptor: | CITRIC ACID, PHOSPHATE ION, Probable diaminopimelate decarboxylase protein | | Authors: | Tang, J, Ju, Y, Zhou, H. | | Deposit date: | 2019-08-05 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
6K7X
 
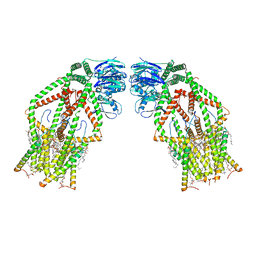 | | Human MCU-EMRE complex | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
