7DA4
 
 | | Cryo-EM structure of amyloid fibril formed by human RIPK3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Zhao, K, Ma, Y.Y, Sun, Y.P, Li, D, Liu, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6L4S
 
 | |
3BIY
 
 | | Crystal structure of p300 histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | BROMIDE ION, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Liu, X, Wang, L, Zhao, K, Thompson, P.R, Hwang, Y, Marmorstein, R, Cole, P.A. | | Deposit date: | 2007-12-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of protein acetylation by the p300/CBP transcriptional coactivator
Nature, 451, 2008
|
|
6LRQ
 
 | |
6LNI
 
 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6L1T
 
 | | Cryo-EM structure of phosphorylated Tyr39 a-synuclein amyloid fibril | | Descriptor: | Alpha-synuclein | | Authors: | Liu, C, Li, Y.M, Zhao, K, Lim, Y.J, Liu, Z.Y. | | Deposit date: | 2019-09-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Parkinson's disease-related phosphorylation at Tyr39 rearranges alpha-synuclein amyloid fibril structure revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L1U
 
 | | Cryo-EM structure of phosphorylated Tyr39 alpha-synuclein amyloid fibril | | Descriptor: | Alpha-synuclein | | Authors: | Liu, C, Li, Y.M, Zhao, K, Lim, Y.J, Liu, Z.Y. | | Deposit date: | 2019-09-30 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Parkinson's disease-related phosphorylation at Tyr39 rearranges alpha-synuclein amyloid fibril structure revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7XJX
 
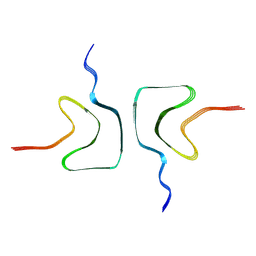 | | The cryo-EM structure of Fe3+ induced alpha-syn fibril. | | Descriptor: | Alpha-synuclein | | Authors: | Zhao, Q.Y, Tao, Y.Q, Zhao, K, Tao, Y.Q, Li, D. | | Deposit date: | 2022-04-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural Insights of Fe3+ Induced alpha-synuclein Fibrillation in Parkinson' Disease
J.Mol.Biol., 435, 2023
|
|
2QQG
 
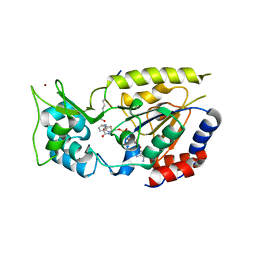 | | Hst2 bound to ADP-HPD, acetyllated histone H4 and nicotinamide | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Histone H4, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B, Zhao, K, Slama, J. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
7BX7
 
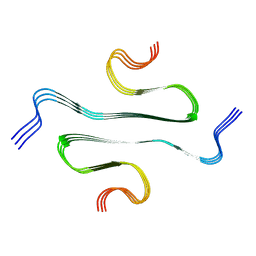 | |
7C1D
 
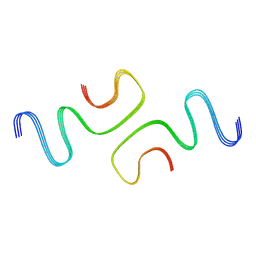 | |
2QQF
 
 | | Hst2 bound to ADP-HPD and Acetylated histone H4 | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Histone H4, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B.D, Zhao, K, Slama, J. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
2YB8
 
 | | Crystal structure of Nurf55 in complex with Su(z)12 | | Descriptor: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
2YBA
 
 | | Crystal structure of Nurf55 in complex with histone H3 | | Descriptor: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
4MZ7
 
 | | Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Zhu, C, Gao, W, Zhao, K, Qin, X, Zhang, Y, Peng, X, Zhang, L, Dong, Y, Zhang, W, Li, P, Wei, W, Gong, Y, Yu, X.F. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase
Nat Commun, 4, 2013
|
|
8IHU
 
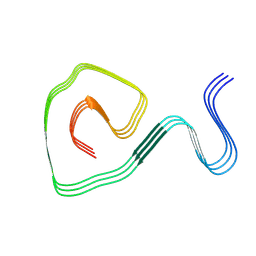 | | Cryo-EM structure of an amyloid fibril formed by ALS-causing SOD1 mutation G85R | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Zhang, M.Y, Yuan, H.Y, Li, X.N, Zhao, K, Chen, J, Li, D, Wang, Z.Z, Le, W.D, Liu, C, Liang, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Amyloid fibril structures and ferroptosis activation induced by ALS-causing SOD1 mutations.
Sci Adv, 2024
|
|
8IHV
 
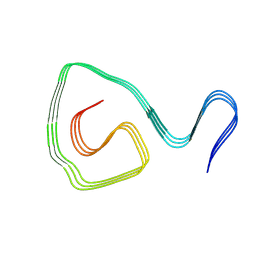 | | Cryo-EM structure of an amyloid fibril formed by ALS-causing SOD1 mutation H46R | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Zhang, M.Y, Yuan, H.Y, Li, X.N, Zhao, K, Chen, J, Li, D, Wang, Z.Z, Le, W.D, Liu, C, Liang, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Amyloid fibril structures and ferroptosis activation induced by ALS-causing SOD1 mutations.
Sci Adv, 2024
|
|
2B9D
 
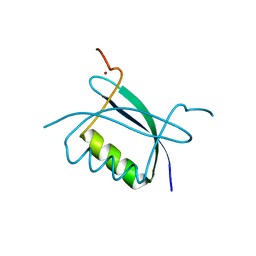 | | Crystal Structure of HPV E7 CR3 domain | | Descriptor: | E7 protein, ZINC ION | | Authors: | Liu, X, Clements, A, Zhao, K, Marmorstein, R. | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor.
J.Biol.Chem., 281, 2006
|
|
1ZX2
 
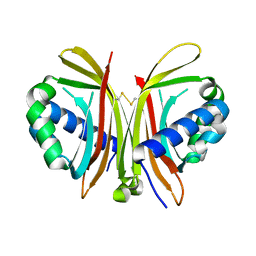 | | Crystal Structure of Yeast UBP3-associated Protein BRE5 | | Descriptor: | UBP3-associated protein BRE5 | | Authors: | Li, K, Zhao, K, Ossareh-Nazari, B, Da, G, Dargemont, C, Marmorstein, R. | | Deposit date: | 2005-06-06 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor
J.Biol.Chem., 280, 2005
|
|
8IVL
 
 | | FABP7 complexed with Cholesterol | | Descriptor: | CHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
8IVF
 
 | | FABP7 complexed with 25-HC | | Descriptor: | 25-HYDROXYCHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
2IOO
 
 | | Crystal structure of the mouse p53 core domain | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IOI
 
 | | Crystal structure of the mouse p53 core domain at 1.55 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
