4J51
 
 | |
8EMR
 
 | | Cryo-EM structure of human liver glucosidase II | | Descriptor: | CALCIUM ION, Glucosidase 2 subunit beta, Neutral alpha-glucosidase AB, ... | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EMS
 
 | | Cryo-EM structure of human liver glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, liver form, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EMT
 
 | | Cryo-EM analysis of the human aldehyde oxidase from liver | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
2YIN
 
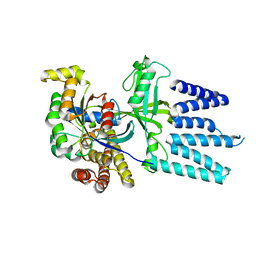 | | STRUCTURE OF THE COMPLEX BETWEEN Dock2 AND Rac1. | | Descriptor: | DEDICATOR OF CYTOKINESIS PROTEIN 2, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 1 | | Authors: | Kulkarni, K.A, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple Factors Confer Specific Cdc42 and Rac Protein Activation by Dedicator of Cytokinesis (Dock) Nucleotide Exchange Factors.
J.Biol.Chem., 286, 2011
|
|
5A31
 
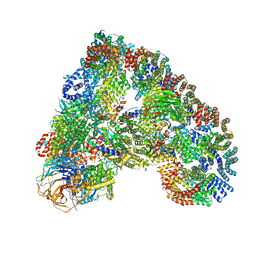 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
4KQE
 
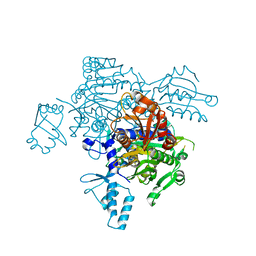 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | Descriptor: | GLYCEROL, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
7Y4F
 
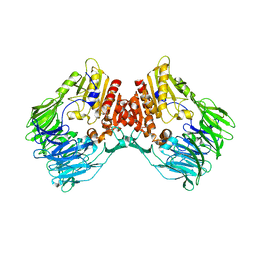 | | bacterial DPP4 | | Descriptor: | Dipeptidyl peptidase IV | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
2E9W
 
 | | Crystal structure of the extracellular domain of Kit in complex with stem cell factor (SCF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Kit ligand, Mast/stem cell growth factor receptor | | Authors: | Yuzawa, S, Opatowsky, Y, Zhang, Z, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2007-01-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Cell(Cambridge,Mass.), 130, 2007
|
|
2EC8
 
 | | Crystal structure of the exctracellular domain of the receptor tyrosine kinase, Kit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mast/stem cell growth factor receptor | | Authors: | Yuzawa, S, Opatowsky, Y, Zhang, Z, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2007-02-11 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Cell(Cambridge,Mass.), 130, 2007
|
|
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
3KAE
 
 | | Cdc27 N-terminus | | Descriptor: | CHLORIDE ION, GLYCEROL, Possible protein of nuclear scaffold, ... | | Authors: | Barford, D, Zhang, Z, Roe, S.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Molecular structure of the N-terminal domain of the APC/C subunit Cdc27 reveals a homo-dimeric tetratricopeptide repeat architecture
J.Mol.Biol., 397, 2010
|
|
1ZCL
 
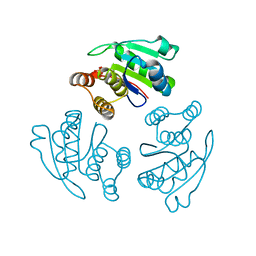 | | prl-1 c104s mutant in complex with sulfate | | Descriptor: | SULFATE ION, protein tyrosine phosphatase 4a1 | | Authors: | Sun, J.P, Wang, W.Q, Yang, H, Liu, S, Liang, F, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2005-04-12 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Biochemical Properties of PRL-1, a Phosphatase Implicated in Cell Growth, Differentiation, and Tumor Invasion.
Biochemistry, 44, 2005
|
|
3J2O
 
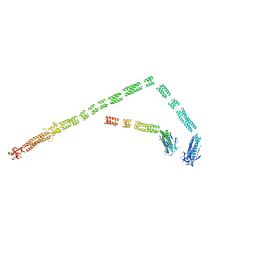 | | Model of the bacteriophage T4 fibritin based on the cryo-EM reconstruction of the contracted T4 tail containing the phage collar and whiskers | | Descriptor: | Fibritin | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A.A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3TN6
 
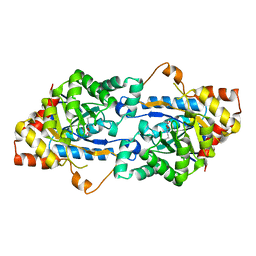 | | Crystal structure of GkaP mutant R230H from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
3TNB
 
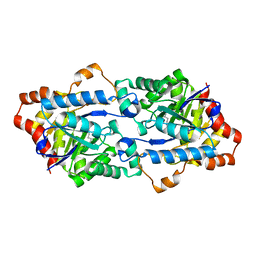 | | Crystal structure of GkaP mutant G209D/R230H from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
1X11
 
 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
1JOT
 
 | | STRUCTURE OF THE LECTIN MPA COMPLEXED WITH T-ANTIGEN DISACCHARIDE | | Descriptor: | AGGLUTININ, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Lee, X, Thompson, A, Zhang, Z, Hoa, T.-T, Biesterfeldt, J, Ogata, C, Xu, L, Johnston, R.A.Z, Young, N.M. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of Maclura pomifera agglutinin and the T-antigen disaccharide, Galbeta1,3GalNAc.
J.Biol.Chem., 273, 1998
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
5ZB9
 
 | | Crystal structure of Rtt109 from Aspergillus fumigatus | | Descriptor: | DNA damage response protein Rtt109, putative, GLYCEROL | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
