7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5C3O
 
 | | Crystal structure of the C-terminal truncated Neurospora crassa T7H (NcT7HdeltaC) in apo form | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Thymine dioxygenase | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C77
 
 | | A novel protein arginine methyltransferase | | Descriptor: | Protein arginine N-methyltransferase SFM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lv, F, Zhang, T, Ding, J. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Sfm1 functioning as a protein arginine methyltransferase.
Cell Discov, 1, 2015
|
|
5XYN
 
 | | The crystal structure of Csm2-Psy3-Shu1-Shu2 complex from budding yeast | | Descriptor: | Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3, Suppressor of HU sensitivity involved in recombination protein 1, ... | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2017-07-09 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the functional role of the Shu complex in homologous recombination.
Nucleic Acids Res., 45, 2017
|
|
8IR0
 
 | | AfFer mutant-P156F | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, C, Zang, J, Zhang, T. | | Deposit date: | 2023-03-17 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Preparation and Unique Three-Dimensional Self-Assembly Property of Starfish Ferritin.
Foods, 12, 2023
|
|
8IQX
 
 | | ferritin mutant-P156H | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, C, Zang, J, Zhang, T. | | Deposit date: | 2023-03-17 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preparation and Unique Three-Dimensional Self-Assembly Property of Starfish Ferritin.
Foods, 12, 2023
|
|
3OV6
 
 | | CD1c in complex with MPM (mannosyl-beta1-phosphomycoketide) | | Descriptor: | 1-O-[(S)-hydroxy{[(4S,8S,16S,20S)-4,8,12,16,20-pentamethylheptacosyl]oxy}phosphoryl]-beta-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Scharf, L, Li, N.S, Hawk, A.J, Garzon, D, Zhang, T, Kazen, A.R, Shah, S, Haddadian, E.J, Saghatelian, A, Faraldo-Gomez, J.D, Meredith, S.C, Piccirilli, J.A, Adams, E.J. | | Deposit date: | 2010-09-15 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation
Immunity, 33, 2010
|
|
4LRV
 
 | | Crystal structure of DndE from Escherichia coli B7A involved in DNA phosphorothioation modification | | Descriptor: | DNA sulfur modification protein DndE | | Authors: | Hu, W, Wang, C.K, Liang, J.D, Zhang, T.L, Yang, M, Hu, Z.P, Wang, Z.J, Lan, W.X, Wu, H.M, Ding, J.P, Wu, G, Deng, Z.X, Cao, C. | | Deposit date: | 2013-07-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification
Cell Res., 22, 2012
|
|
5WQA
 
 | | Crystal structure of PDE4D catalytic domain complexed with Selaginpulvilins K | | Descriptor: | 1-[2-(4-hydroxyphenyl)ethynyl]-9,9-bis(4-methoxyphenyl)-7-oxidanyl-fluorene-2-carbaldehyde, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y, Zhang, T, Zheng, X, Yin, S, Luo, H.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The discovery, complex crystal structure, and recognition mechanism of a novel natural PDE4 inhibitor from Selaginella pulvinata
Biochem. Pharmacol., 130, 2017
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6.343 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
7BTA
 
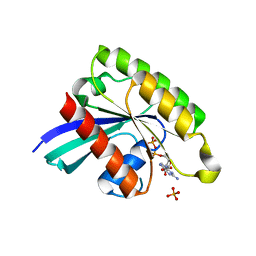 | | Crystal structure of Rheb D60K mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-03-31 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTC
 
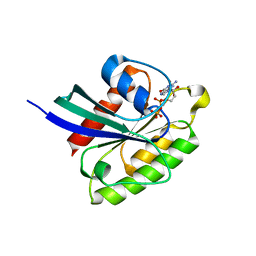 | | Crystal structure of Rheb Y35N mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTD
 
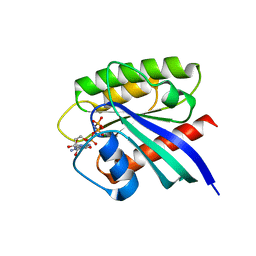 | | Crystal structure of Rheb Y35N mutant bound to GppNHp | | Descriptor: | GTP-binding protein Rheb, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
5Y6M
 
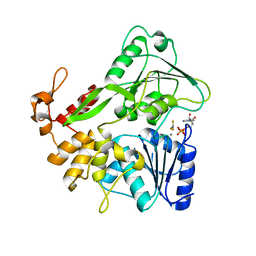 | | Zika virus helicase in complex with ADP-AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
3RIB
 
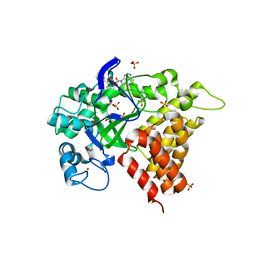 | | Human lysine methyltransferase Smyd2 in complex with AdoHcy | | Descriptor: | N-lysine methyltransferase SMYD2, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Xu, S, Zhang, T, Zhong, C, Ding, J. | | Deposit date: | 2011-04-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of human lysine methyltransferase Smyd2 reveals insights into the substrate divergence in Smyd proteins
J Mol Cell Biol, 3, 2011
|
|
5Y6N
 
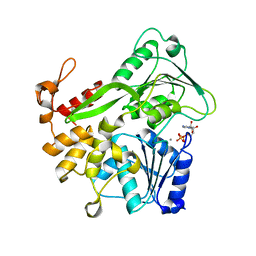 | | Zika virus helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase domain from Genome polyprotein, MANGANESE (II) ION | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
4M6W
 
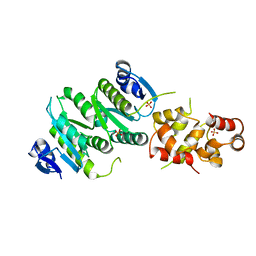 | | Crystal structure of the C-terminal segment of FANCM in complex with FAAP24 | | Descriptor: | Fanconi anemia group M protein, Fanconi anemia-associated protein of 24 kDa, SULFATE ION | | Authors: | Yang, H, Zhang, T, Tong, L, Ding, J. | | Deposit date: | 2013-08-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair.
Nucleic Acids Res., 41, 2013
|
|
4LTU
 
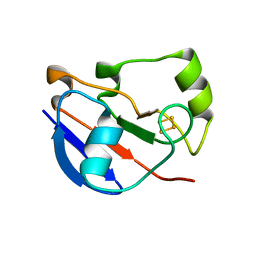 | | Crystal Structure of Ferredoxin from Rhodopseudomonas palustris HaA2 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Zhou, W.H, Zhang, T, Yang, H, Bell, S.G, Wong, L.-L. | | Deposit date: | 2013-07-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Ferredoxin from Rhodopseudomonas palustris HaA2
To be Published
|
|
6KEA
 
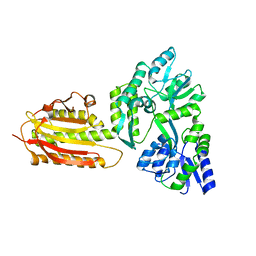 | | crystal structure of MBP-tagged REV7-IpaB complex | | Descriptor: | Maltose-binding periplasmic protein,LINKER,hREV7,LINKER,Invasin IpaB,hREV3 | | Authors: | Wang, X, Pernicone, N, Pertz, L, Hua, D.P, Zhang, T.Q, Listovsky, T, Xie, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | REV7 has a dynamic adaptor region to accommodate small GTPase RAN/ShigellaIpaB ligands, and its activity is regulated by the RanGTP/GDP switch.
J.Biol.Chem., 294, 2019
|
|
4FBG
 
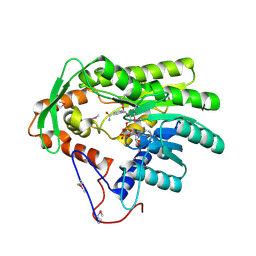 | | Crystal structure of Treponema denticola trans-2-enoyl-CoA reductase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative reductase TDE_0597 | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-05-23 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
3RPN
 
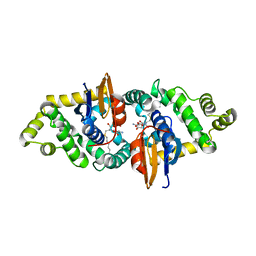 | | Crystal structure of human kappa class glutathione transferase in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase kappa 1, S-HEXYLGLUTATHIONE | | Authors: | Wang, B, Peng, Y, Zhang, T, Ding, J. | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and kinetic studies of human Kappa class glutathione transferase provide insights into the catalytic mechanism.
Biochem.J., 439, 2011
|
|
3RPP
 
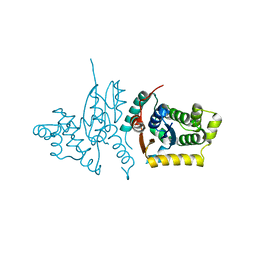 | |
6V97
 
 | | Kindlin-3 double deletion mutant short form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
