8HGV
 
 | |
5C6D
 
 | | Crystal structure of USP7 in complex with UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Zhang, Z.-M, Song, J. | | Deposit date: | 2015-06-22 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1.
Cell Rep, 12, 2015
|
|
4LJY
 
 | | Crystal structure of RNA splicing effector Prp5 in complex with ADP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-12-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
8K3J
 
 | | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2, ... | | Authors: | Zhang, Z.Z. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer
To Be Published
|
|
1W9Y
 
 | | The structure of ACC oxidase | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE 1, SULFATE ION | | Authors: | Zhang, Z, Ren, J.-S, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2004-10-20 | | Release date: | 2005-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanistic Implications of 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (the Ethyling Forming Enzyme)
Chem.Biol., 11, 2004
|
|
5H4I
 
 | |
1WA6
 
 | | The structure of ACC oxidase | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE 1, FE (II) ION, PHOSPHATE ION, ... | | Authors: | Zhang, Z, Ren, J.-S, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2004-10-25 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure and Mechanistic Implications of 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (the Ethyling Forming Enzyme)
Chem.Biol., 11, 2004
|
|
3G0C
 
 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinedione inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(2-chlorobenzyl)-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-1H-purine-2,6-dione, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
8FFK
 
 | |
8FFS
 
 | |
6Y50
 
 | | 5'domain of human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | Descriptor: | DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
6WGN
 
 | | Crystal structure of K-Ras(G12D) GppNHp bound to cyclic peptide ligand KD2 | | Descriptor: | Cyclic Peptide KD2, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Gao, R, Hu, Q, Peacock, H, Peacock, D.M, Shokat, K.M, Suga, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Acs Cent.Sci., 6, 2020
|
|
8K1W
 
 | |
4GE6
 
 | |
6LW1
 
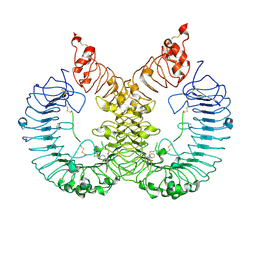 | | Cryo-EM structure of TLR7/Cpd-7 (DSR-139970) complex in open form | | Descriptor: | 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-6-methyl-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purine, Toll-like receptor 7 | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
5ZSC
 
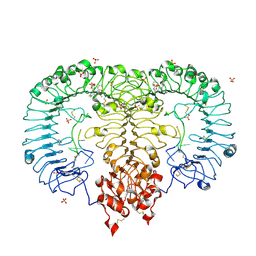 | | Crystal structure of monkey TLR7 in complex with IMDQ and CCUUCC | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
2QMY
 
 | | Quinone Reductase 2 in complex with adrenochrome | | Descriptor: | (3S)-3-hydroxy-1-methyl-2,3-dihydro-1H-indole-5,6-dione, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase, ... | | Authors: | Zhang, Z, Fu, Y, Buryanovskky, L. | | Deposit date: | 2007-07-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quinone Reductase 2 regulates Catecholamine oxidation
To be Published
|
|
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2013-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
7E3J
 
 | |
6K10
 
 | | Non substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78962183 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
8UX2
 
 | | Chromobacterium violaceum mono-ADP-ribosyltransferase CteC in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NAD(+)--protein-threonine ADP-ribosyltransferase, ... | | Authors: | Zhang, Z, Rondon, H, Das, C. | | Deposit date: | 2023-11-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of bacterial ubiquitin ADP-ribosyltransferase CteC reveals a substrate-recruiting insertion.
J.Biol.Chem., 300, 2023
|
|
8QP9
 
 | | Cryo-EM Structure of Pre-B+AMPPNP Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QP8
 
 | | Cryo-EM Structure of Pre-B Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPE
 
 | | Cryo-EM Structure of Pre-B-like Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
