178L
 
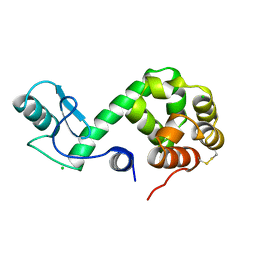 | | Protein flexibility and adaptability seen in 25 crystal forms of T4 LYSOZYME | | 分子名称: | CHLORIDE ION, T4 LYSOZYME | | 著者 | Matsumura, M, Weaver, L, Zhang, X.-J, Matthews, B.W. | | 登録日 | 1995-03-24 | | 公開日 | 1995-07-10 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | 分子名称: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | 著者 | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | 登録日 | 2023-09-26 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (6.04 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
3J6C
 
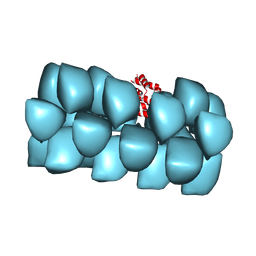 | | Cryo-EM structure of MAVS CARD filament | | 分子名称: | Mitochondrial antiviral-signaling protein | | 著者 | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | 登録日 | 2014-02-04 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.6 Å) | | 主引用文献 | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
167L
 
 | |
180L
 
 | |
6NT7
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound dimeric state | | 分子名称: | Stimulator of interferon genes protein, cGAMP | | 著者 | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | 登録日 | 2019-01-28 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
5CMS
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, SULFATE ION | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-07-17 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
6GFW
 
 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Zhang, X.D. | | 登録日 | 2018-05-02 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
3UT4
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | 分子名称: | Putative uncharacterized protein | | 著者 | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | 登録日 | 2011-11-25 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
2VII
 
 | | PspF1-275-Mg-AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | 著者 | Joly, N, Rappas, M, Buck, M, Zhang, X. | | 登録日 | 2007-12-04 | | 公開日 | 2008-01-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Trapping of a Transcription Complex Using a New Nucleotide Analogue: AMP Aluminium Fluoride
J.Mol.Biol., 375, 2008
|
|
6GH6
 
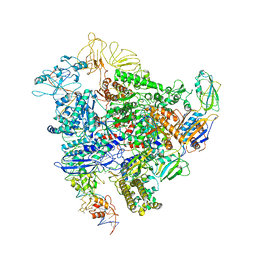 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme intermediate partially loaded complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Zhang, X.D. | | 登録日 | 2018-05-04 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
3CI9
 
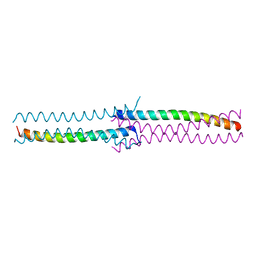 | | Crystal Structure of the human HSBP1 | | 分子名称: | Heat shock factor-binding protein 1 | | 著者 | Liu, X, Xu, L, Liu, Y, Zhu, G, Zhang, X.C, Li, X, Rao, Z. | | 登録日 | 2008-03-11 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the hexamer of human heat shock factor binding protein 1
Proteins, 75, 2009
|
|
1MVX
 
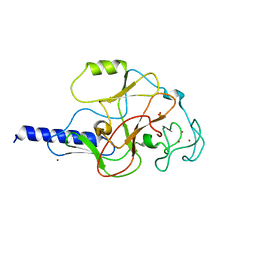 | | structure of the SET domain histone lysine methyltransferase Clr4 | | 分子名称: | CRYPTIC LOCI REGULATOR 4, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | 登録日 | 2002-09-26 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
1MVH
 
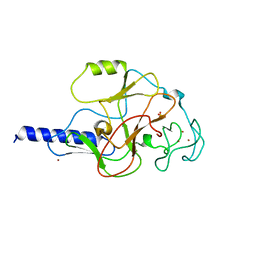 | | structure of the SET domain histone lysine methyltransferase Clr4 | | 分子名称: | Cryptic loci regulator 4, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | 登録日 | 2002-09-25 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | 分子名称: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | 著者 | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2015-11-25 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
7YMI
 
 | | PSII-Pcb Dimer of Acaryochloris Marina | | 分子名称: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | 著者 | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | 登録日 | 2022-07-28 | | 公開日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
5EH2
 
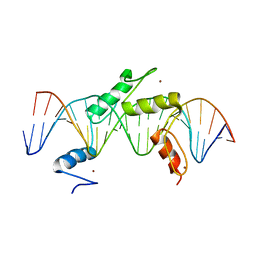 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence III | | 分子名称: | DNA (5'-D(*AP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | 著者 | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | 登録日 | 2015-10-27 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EI9
 
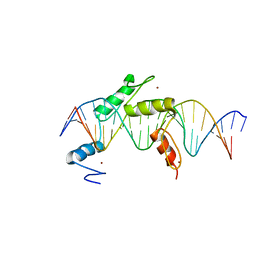 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence I | | 分子名称: | DNA (5'-D(*AP*TP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*GP*A)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | 著者 | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | 登録日 | 2015-10-29 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.921 Å) | | 主引用文献 | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
6IYA
 
 | |
6W38
 
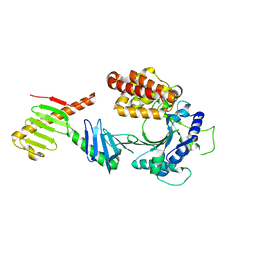 | | Crystal structure of the FAM46C/Plk4 complex | | 分子名称: | Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | 著者 | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (4.48 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
1C76
 
 | | STAPHYLOKINASE (SAK) MONOMER | | 分子名称: | STAPHYLOKINASE | | 著者 | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | 登録日 | 2000-02-01 | | 公開日 | 2000-08-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of Staphylokinase Dimer Offers New Clue to Reduction of Immunogenicity
To be published
|
|
8II0
 
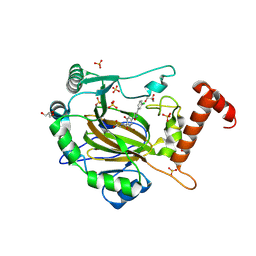 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(3-(3-chlorophenyl)isoxazol-5-yl)-3-hydroxypicolinoyl)glycine | | 分子名称: | 2-[[5-[3-(3-chlorophenyl)-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | 著者 | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | 登録日 | 2023-02-24 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8IHZ
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(1-(3-(4-chlorophenyl)propyl)-1H-1,2,3-triazol-4-yl)-3-hydroxypicolinoyl)glycine | | 分子名称: | 2-[[5-[1-[3-(4-chlorophenyl)propyl]-1,2,3-triazol-4-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | 著者 | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | 登録日 | 2023-02-24 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 2024
|
|
5COA
 
 | | Crystal structure of iridoid synthase at 2.2-angstrom resolution | | 分子名称: | HEXAETHYLENE GLYCOL, Iridoid synthase, SULFATE ION | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
