7EDA
 
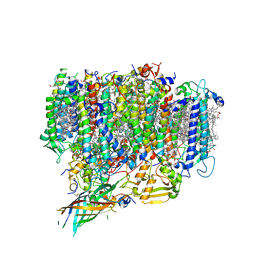 | | Structure of monomeric photosystem II | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Yu, H, Hamaguchi, T, Nakajima, Y, Kato, K, kawakami, K, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Cryo-EM structure of monomeric photosystem II at 2.78 angstrom resolution reveals factors important for the formation of dimer.
Biochim Biophys Acta Bioenerg, 1862, 2021
|
|
7CC9
 
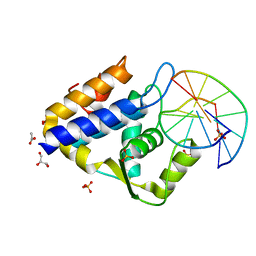 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*GP*CP*GP*GS*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*CP*GP*CP*C)-3'), ... | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7CCJ
 
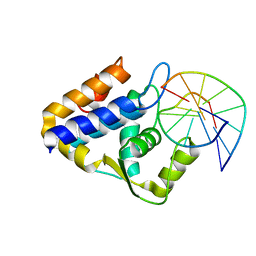 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*AP*TP*C)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7CCD
 
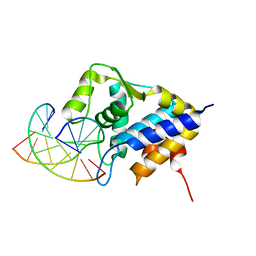 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*GP*AS*AP*CP*GP*TP*G)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Li, J, Liu, G, Zhao, G, Wang, Y, Hu, W, Deng, Z, Gan, J, Zhao, Y, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
6WIV
 
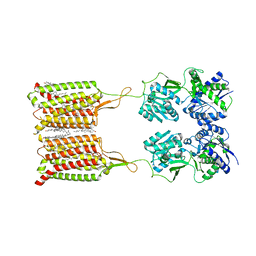 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
2UXN
 
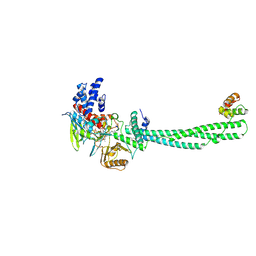 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | Descriptor: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2UXX
 
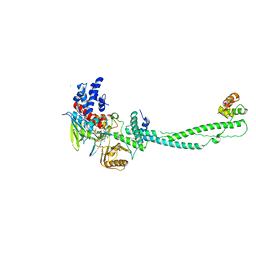 | | Human LSD1 Histone Demethylase-CoREST in complex with an FAD- tranylcypromine adduct | | Descriptor: | CHLORIDE ION, FAD-trans-2-Phenylcyclopropylamine Adduct, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for the Inhibition of the Lsd1 Histone Demethylase by the Antidepressant Trans-2-Phenylcyclopropylamine.
Biochemistry, 46, 2007
|
|
1S2H
 
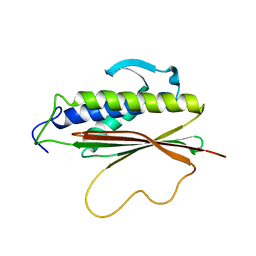 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RLQ
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1RLP
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRM
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRL
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
5KGN
 
 | | 1.95A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (2d) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGM
 
 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGL
 
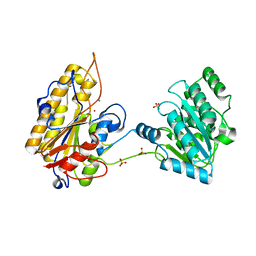 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
6WG3
 
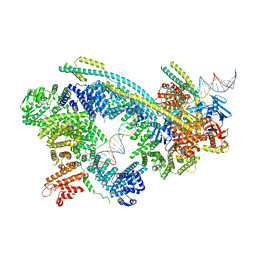 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex | | Descriptor: | Cohesin subunit SA-1, DNA (51-MER), Double-strand-break repair protein rad21 homolog, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
6WGE
 
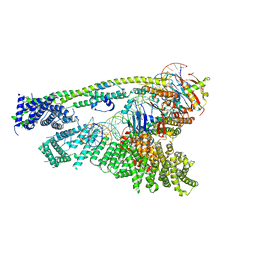 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex without STAG1 | | Descriptor: | DNA (43-MER), Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
3SHU
 
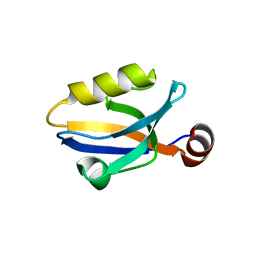 | | Crystal structure of ZO-1 PDZ3 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
6WG6
 
 | |
6MDW
 
 | | Mechanism of protease dependent DPC repair | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CITRATE ANION, ... | | Authors: | Li, F, Raczynska, J, Chen, Z, Yu, H. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into DNA-Dependent Activation of Human Metalloprotease Spartan.
Cell Rep, 26, 2019
|
|
6WG4
 
 | |
6WSL
 
 | | Cryo-EM structure of VASH1-SVBP bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Small vasohibin-binding protein, ... | | Authors: | Li, F, Li, Y, Yu, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of VASH1-SVBP bound to microtubules.
Elife, 9, 2020
|
|
6BNT
 
 | |
1PKS
 
 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
1PKT
 
 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
