5TFA
 
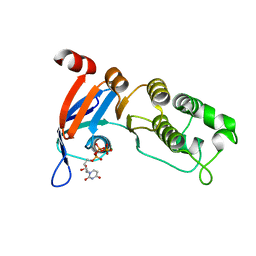 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dUTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DEOXYURIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFG
 
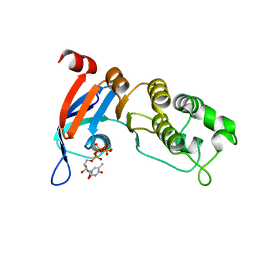 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with 5-methyl-UTP | | Descriptor: | 5-Methyluridine triphosphate, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
3N71
 
 | |
6IH0
 
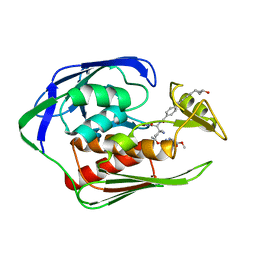 | | Aquifex aeolicus LpxC complex with ACHN-975 | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Li, D.Y, Fan, S, Jin, Y.Y, Zhang, C, Lv, G.X, Wu, G.T, Yang, Z.Y. | | Deposit date: | 2018-09-27 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Complex Structure of Protein AaLpxC from Aquifex aeolicus with ACHN-975 Molecule Suggests an Inhibitory Mechanism at Atomic-Level against Gram-Negative Bacteria
Molecules, 26, 2021
|
|
4P0C
 
 | | Crystal Structure of NHERF2 PDZ1 Domain in Complex with LPA2 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF2/Lysophosphatidic acid receptor 2 chimeric protein, THIOCYANATE ION | | Authors: | Holcomb, J, Jiang, Y, Lu, G, Trescott, L, Brunzelle, J, Sirinupong, N, Li, C, Naren, A, Yang, Z. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural insights into PDZ-mediated interaction of NHERF2 and LPA2, a cellular event implicated in CFTR channel regulation.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4O6F
 
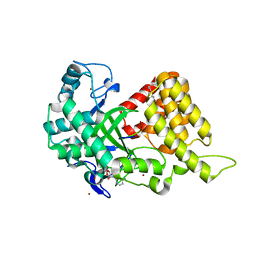 | | Structural Basis of Estrogen Receptor Alpha Methylation Mediated by Histone Methyltransferase SmyD2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Estrogen receptor, N-lysine methyltransferase SMYD2, ... | | Authors: | Jiang, Y, Trescott, L, Holcomb, J, Brunzelle, J, Sirinupong, N, Shi, X, Yang, Z. | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Structural Insights into Estrogen Receptor alpha Methylation by Histone Methyltransferase SMYD2, a Cellular Event Implicated in Estrogen Signaling Regulation.
J.Mol.Biol., 426, 2014
|
|
4PQW
 
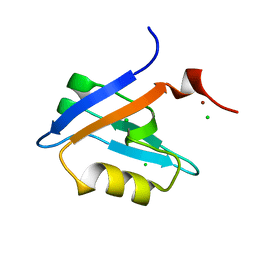 | | Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Wang, S, Holcomb, J, Trescott, L, Guan, X, Hou, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic analysis of NHERF1-PLC beta 3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4Q3H
 
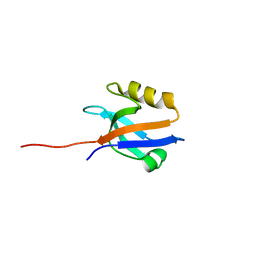 | | The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Holcomb, J, Jiang, Y, Trescott, L, Lu, G, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization.
Biochem.Biophys.Res.Commun., 448, 2014
|
|
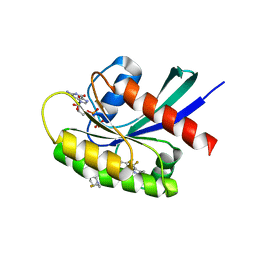
8TXG
 
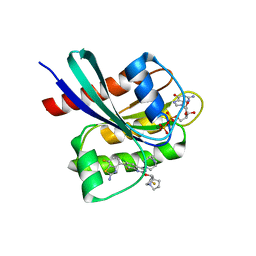 | | Crystal structure of KRAS G12D in complex with GDP and compound 8 | | Descriptor: | (4M)-4-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-7-fluoro-1,3-benzothiazol-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8TXE
 
 | | Crystal structure of KRAS G12D in complex with GDP and compound 5 | | Descriptor: | (6M)-6-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-4-methyl-5-(trifluoromethyl)pyridin-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8TXH
 
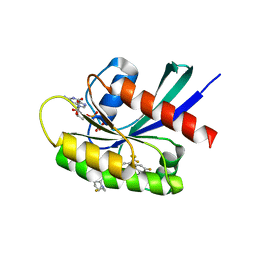 | | Crystal structure of KRAS G12D in complex with GDP and compound 14 | | Descriptor: | (4P)-2-amino-4-{4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}-6-(trifluoromethyl)quinazolin-7-yl}-7-fluoro-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
4JL7
 
 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-03-12 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Insights into Neutrophilic Migration Revealed by the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1.
Plos One, 8, 2013
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4N6X
 
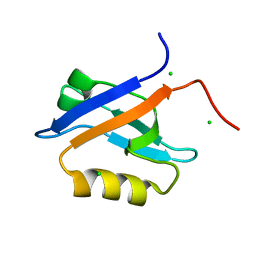 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1/Chemokine Receptor CXCR2 fusion protein | | Authors: | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.051 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
7XHO
 
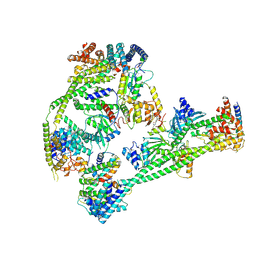 | | Structure of human inner kinetochore CCAN complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Tian, T, Wang, C.L, Yang, Z.S, Sun, L.F, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
7XHN
 
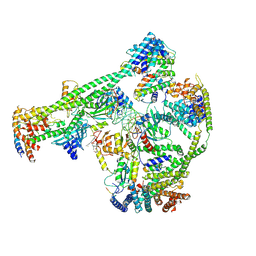 | | Structure of human inner kinetochore CCAN-DNA complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Sun, L.F, Tian, T, Wang, C.L, Yang, Z.S, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
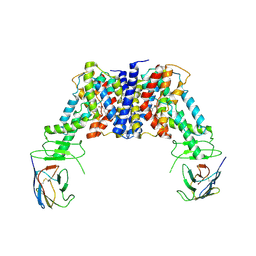
8JH5
 
 | |
7UW4
 
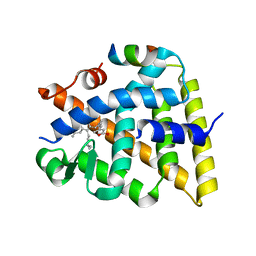 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB113 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-propylcyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
4Z8U
 
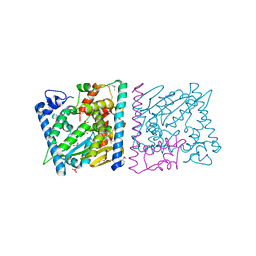 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:-ORF2 WITH ATP | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8Q
 
 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 COMPLEX, SELENOMETHIONINE SUBSTITUTED. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
7EER
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with 05E6 and ATP | | Descriptor: | 2-(1H-indol-3-yl)ethanol, ADENOSINE-5'-TRIPHOSPHATE, Tryptophan--tRNA ligase | | Authors: | Lv, G, Fan, S, Feng, X, Zhang, Q, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2021-03-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with O5E6 and ATP
To Be Published
|
|
4Z8V
 
 | | CRYSTAL STRUCTURE OF AVRRXO1-ORF1:-ORF2 COMPLEX, NATIVE. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
