5BPW
 
 | |
5BPZ
 
 | | Atomic-resolution structures of the APC/C subunits Apc4 and the Apc5 N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Anapc5 protein | | Authors: | Cronin, N, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2015-05-28 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Atomic-Resolution Structures of the APC/C Subunits Apc4 and the Apc5 N-Terminal Domain.
J.Mol.Biol., 427, 2015
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
1UAK
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAL
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
7K9P
 
 | | Room temperature structure of NSP15 Endoribonuclease from SARS CoV-2 solved using SFX. | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Botha, S, Jernigan, R, Chen, J, Coleman, M.A, Frank, M, Grant, T.D, Hansen, D.T, Ketawala, G, Logeswaran, D, Martin-Garcia, J, Nagaratnam, N, Raj, A.L.L.X, Shelby, M, Yang, J.-H, Yung, M.C, Fromme, P. | | Deposit date: | 2020-09-29 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Room-temperature structural studies of SARS-CoV-2 protein NendoU with an X-ray free-electron laser.
Structure, 2022
|
|
8X6M
 
 | | Crystal Structure of Glycerol Dehydrogenase in the Presence of NAD+ and Glycerol | | Descriptor: | GLYCEROL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Kang, J.Y, Jin, M, Yang, J, Kim, H, Noh, C, Eom, S.H. | | Deposit date: | 2023-11-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the octamerization of glycerol dehydrogenase.
Plos One, 19, 2024
|
|
5B6C
 
 | | Structural Details of Ufd1 binding to p97 | | Descriptor: | Peptide from Ubiquitin fusion degradation protein 1 homolog, Transitional endoplasmic reticulum ATPase | | Authors: | Le, L.T.M, Yang, J.K. | | Deposit date: | 2016-05-26 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation
PLoS ONE, 11, 2016
|
|
5GLF
 
 | | Structural insights into the interaction of p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease | | Descriptor: | Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Yoon, S.Y, Ly, T.T, Kang, J.Y, Youn, H.-S, An, J.Y, Lee, J.-G, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2016-07-11 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the interaction of human p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease.
FEBS Lett., 590, 2016
|
|
5EPP
 
 | | Structural Insights into the Interaction of p97 N-terminus Domain and VBM Motif in Rhomboid Protease, RHBDL4 | | Descriptor: | Rhomboid-related protein 4, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Ly, T.T, Kang, J.Y, Lee, J.-G, An, J.Y, Youn, H.-S, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2015-11-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into the interaction of p97 N-terminus domain and VBM in rhomboid protease, RHBDL4.
Biochem.J., 473, 2016
|
|
7CE8
 
 | | Crystal structure of T2R-TTL-Compound11 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5A31
 
 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2IGG
 
 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
3PVB
 
 | | Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Boettcher, A.J, Wu, J, Kim, C, Yang, J, Bruystens, J, Cheung, N, Pennypacker, J.K, Blumenthal, D.A, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2010-12-06 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase
Structure, 19, 2011
|
|
1UW0
 
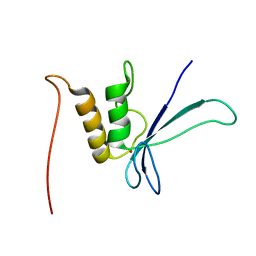 | |
2L5G
 
 | | Co-ordinates and 1H, 13C and 15N chemical shift assignments for the complex of GPS2 53-90 and SMRT 167-207 | | Descriptor: | G protein pathway suppressor 2, Putative uncharacterized protein NCOR2 | | Authors: | Oberoi, J, Yang, J, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1SYK
 
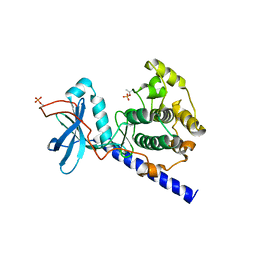 | | Crystal structure of E230Q mutant of cAMP-dependent protein kinase reveals unexpected apoenzyme conformation | | Descriptor: | cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Wu, J, Yang, J, Madhusudan, N, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the E230Q mutant of cAMP-dependent
protein kinase reveals an unexpected apoenzyme conformation and an
extended N-terminal A helix.
Protein Sci., 14, 2005
|
|
2JMX
 
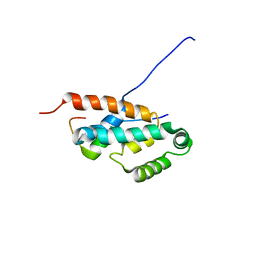 | | OSCP-NT (1-120) in complex with N-terminal (1-25) alpha subunit from F1-ATPase | | Descriptor: | ATP synthase O subunit, mitochondrial, ATP synthase subunit alpha heart isoform | | Authors: | Carbajo, R.J, Neuhaus, D, Kellas, F.A, Yang, J, Runswick, M.J, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | How the N-terminal Domain of the OSCP Subunit of Bovine F(1)F(o)-ATP Synthase Interacts with the N-terminal Region of an Alpha Subunit
J.Mol.Biol., 368, 2007
|
|
7AAC
 
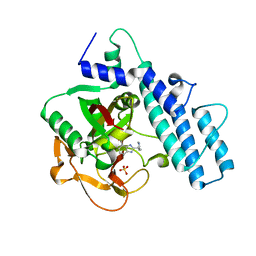 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAD
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
6J0N
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, baseplate in extended state, refined in C6 symmetry | | Descriptor: | Pvc1, Pvc11, Pvc12, ... | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
4QLA
 
 | | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori | | Descriptor: | Juvenile hormone epoxide hydrolase, PENTAETHYLENE GLYCOL | | Authors: | Zhou, K, Jia, N, Hu, C, Jiang, Y.L, Yang, J.P, Chen, Y, Li, S, Zhou, C.Z. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori.
Proteins, 82, 2014
|
|
