6KGW
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
5GVV
 
 | | Crystal structure of the glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
5GVW
 
 | | Crystal structure of the apo-form glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
3KYS
 
 | | Crystal structure of human YAP and TEAD complex | | Descriptor: | 65 kDa Yes-associated protein, Transcriptional enhancer factor TEF-1 | | Authors: | Li, Z, Zhao, B, Wang, P, Chen, F, Dong, Z, Yang, H, Guan, K.L, Xu, Y. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-23 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the YAP and TEAD complex
Genes Dev., 24, 2010
|
|
4UBD
 
 | | Crystal structure of a neutralizing human monoclonal antibody with 1968 H3 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Shore, D.A, Yang, H, Cho, M, Donis, R.O, Stevens, J. | | Deposit date: | 2014-08-12 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus.
Nat Commun, 6, 2015
|
|
4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.916 Å) | | Cite: | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
5H2A
 
 | |
5H28
 
 | |
7CKF
 
 | | The N-terminus of interferon-inducible antiviral protein-dimer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Yang, H.T. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUT
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with Z-VAD-FMK | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7CUU
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with MG132 | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
4US3
 
 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
4US4
 
 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | Authors: | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7XYF
 
 | |
7XYG
 
 | |
7C8H
 
 | | Ambient temperature structure of Bifidobacterium longum phosphoketolase with thiamine diphosphate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, MALONIC ACID, ... | | Authors: | Nakata, K, Kashiwagi, T, Nango, E, Miyano, H, Mizukoshi, T, Iwata, S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ambient temperature structure of phosphoketolase from Bifidobacterium longum determined by serial femtosecond X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7C8I
 
 | | Ambient temperature structure of Bifidobacgterium longum phosphoketolase with thiamine diphosphate and phosphoenol pyuruvate | | Descriptor: | CALCIUM ION, PHOSPHOENOLPYRUVATE, THIAMINE DIPHOSPHATE, ... | | Authors: | Nakata, K, Kashiwagi, T, Nango, E, Miyano, H, Mizukoshi, T, Iwata, S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ambient temperature structure of phosphoketolase from Bifidobacterium longum determined by serial femtosecond X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8KFO
 
 | | Crystal structure of BSA in complex with B3 | | Descriptor: | 6-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]-~{N},~{N}-dimethyl-naphthalen-2-amine, Albumin | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Bioinspired two-stage assembled photosensitive protein engineering for tumor-specific mitochondrial targeted phototherapy
To Be Published
|
|
7PD3
 
 | | Structure of the human mitoribosomal large subunit in complex with NSUN4.MTERF4.GTPBP7 and MALSU1.L0R8F8.mt-ACP | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Chandrasekaran, V, Desai, N, Burton, N.O, Yang, H, Price, J, Miska, E.A, Ramakrishnan, V. | | Deposit date: | 2021-08-04 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualizing formation of the active site in the mitochondrial ribosome.
Elife, 10, 2021
|
|
5C94
 
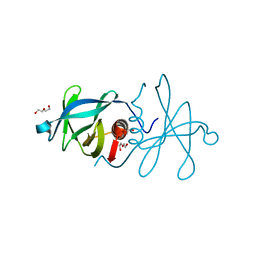 | | Infectious bronchitis virus nsp9 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-structural protein 9 | | Authors: | Chen, C, Dou, Y, Yang, H, Su, D. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9.
Protein Sci., 26, 2017
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
1LB2
 
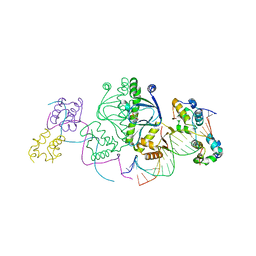 | | Structure of the E. coli alpha C-terminal domain of RNA polymerase in complex with CAP and DNA | | Descriptor: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*AP*G)-3', 5'-D(*CP*TP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Benoff, B, Yang, H, Lawson, C.L, Parkinson, G, Liu, J, Blatter, E, Ebright, Y.W, Berman, H.M, Ebright, R.H. | | Deposit date: | 2002-04-01 | | Release date: | 2002-09-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription activation: the CAP-alpha CTD-DNA complex.
Science, 297, 2002
|
|
1IYJ
 
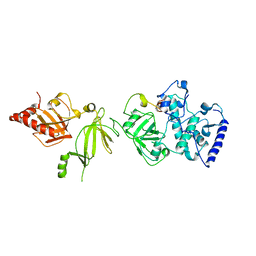 | | STRUCTURE OF A BRCA2-DSS1 COMPLEX | | Descriptor: | Deleted in split hand/split foot protein 1, breast cancer susceptibility | | Authors: | Pavletich, N.P, Jeffrey, P.D, Yang, H.J. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
