6IDF
 
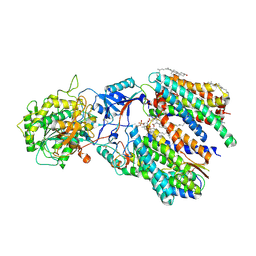 | | Cryo-EM structure of gamma secretase in complex with a Notch fragment | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Zhou, Q, Guo, X, Yan, C, Ke, M, Lei, J, Shi, Y. | | Deposit date: | 2018-09-09 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of Notch recognition by human gamma-secretase
Nature, 565, 2019
|
|
7C9I
 
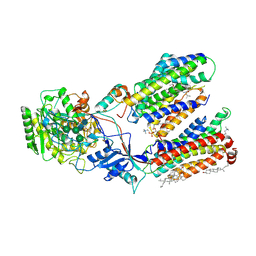 | | Human gamma-secretase in complex with small molecule L-685,458 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7D8X
 
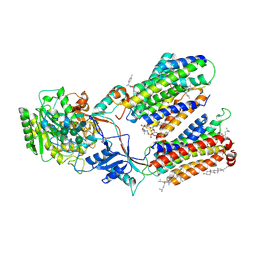 | | CryoEM structure of human gamma-secretase in complex with E2012 and L685458 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[(1S)-1-(4-fluorophenyl)ethyl]-3-[[3-methoxy-4-(4-methylimidazol-1-yl)phenyl]methylidene]piperidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
8HOW
 
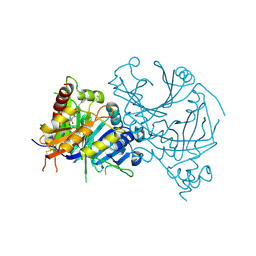 | | Crystal structure of AtHPPD-Y191052 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(2-phenylethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-12-11 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
5AEL
 
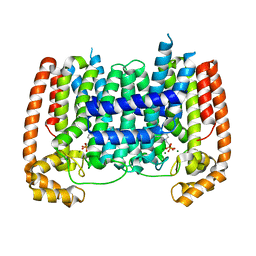 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-597 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, MAGNESIUM ION, {2-[3-(hex-1-yn-1-yl)pyridinium-1-yl]ethane-1,1-diyl}bis(phosphonate) | | Authors: | Yang, G, Oldfield, E, No, J.H. | | Deposit date: | 2014-12-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition of Trypanosoma Brucei Cell Growth by Lipophilic Bisphosphonates: An in Vitro and in Vivo Investigation.
Antimicrob.Agents Chemother., 59, 2015
|
|
5AFX
 
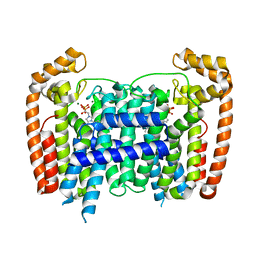 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-1238 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, MAGNESIUM ION, [1-hydroxy-2-(1-nonyl-1H-3lambda~5~-imidazol-3-yl)ethane-1,1-diyl]bis(phosphonic acid) | | Authors: | Yang, G, Oldfield, E, No, J.H. | | Deposit date: | 2015-01-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Inhibition of Trypanosoma Brucei Cell Growth by Lipophilic Bisphosphonates: An in Vitro and in Vivo Investigation.
Antimicrob.Agents Chemother., 59, 2015
|
|
5AHU
 
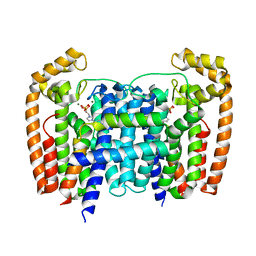 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-1326 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, PUTATIVE, MAGNESIUM ION, ... | | Authors: | Yang, G, Oldfield, E, No, J.H. | | Deposit date: | 2015-02-09 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Inhibition of Trypanosoma Brucei Cell Growth by Lipophilic Bisphosphonates: An in Vitro and in Vivo Investigation.
Antimicrob.Agents Chemother., 59, 2015
|
|
5XGK
 
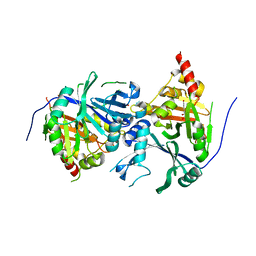 | | Crystal structure of Arabidopsis thaliana 4-hydroxyphenylpyruvate dioxygenase (AtHPPD) complexed with its substrate 4-hydroxyphenylpyruvate acid (HPPA) | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, 4-hydroxyphenylpyruvate dioxygenase, ACETATE ION, ... | | Authors: | Yang, G.F, Yang, W.C, Lin, H.Y. | | Deposit date: | 2017-04-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Res, 2019, 2019
|
|
8GWD
 
 | | Crystal structure of AtHPPD-Y18734 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-[(1S)-1-phenylethyl]quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Crystal structure of AtHPPD-Y18734 complex
To Be Published
|
|
7VO8
 
 | | Structure of AtHPPD complexed with LSY-1 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, N-[6-[[9-(dimethylamino)-12H-benzo[a]phenoxazin-5-yl]amino]hexyl]-4-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-[oxidanyl(oxidanylidene)-$l^4-azanyl]benzamide | | Authors: | Yang, G.-F, Dong, J. | | Deposit date: | 2021-10-12 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In vivo diagnostics of abiotic plant stress responses via in situ real-time fluorescence imaging
Plant Physiol., 190, 2022
|
|
8H6B
 
 | | Crystal structure of AtHPPD complexed with YH20702 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, N-(1-methyl-1,2,3,4-tetrazol-5-yl)-1-pentyl-pyrrolo[2,3-b]pyridine-4-carboxamide | | Authors: | Yang, G.-F. | | Deposit date: | 2022-10-16 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Crystal structure of AtHPPD complexed with YH20702
To Be Published
|
|
8H6A
 
 | | Crystal structure of AtHPPD complexed with YH21477 | | Descriptor: | 1-[(2-chlorophenyl)methyl]-3-methyl-N-(1-methyl-1,2,3,4-tetrazol-5-yl)-2-oxidanylidene-benzimidazole-4-carboxamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F. | | Deposit date: | 2022-10-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of AtHPPD complexed with YH20702
To Be Published
|
|
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JDA
 
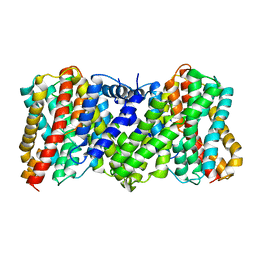 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | Descriptor: | Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
7WSW
 
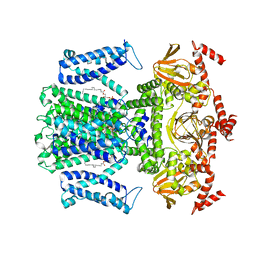 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
8XC3
 
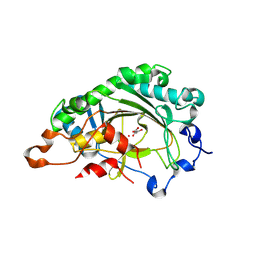 | | Crystal structure of ZmHSL1A-MBQ complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 2-OXOGLUTARIC ACID, 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, ... | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7VC8
 
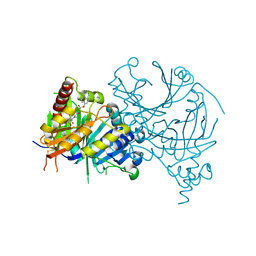 | | Complex structure of AtHPPD with inhibitor PYQ3 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, pyren-1-yl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate | | Authors: | Yang, G.F, Lin, H.Y. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
6LR4
 
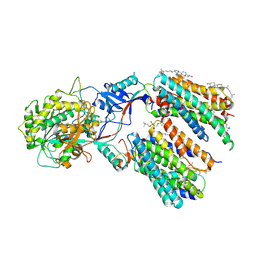 | | Molecular basis for inhibition of human gamma-secretase by small molecule | | Descriptor: | (2S)-2-hydroxy-3-methyl-N-[(2S)-1-[[(5S)-3-methyl-4-oxo-2,5-dihydro-1H-3-benzazepin-5-yl]amino]-1-oxopropan-2-yl]butanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7XUF
 
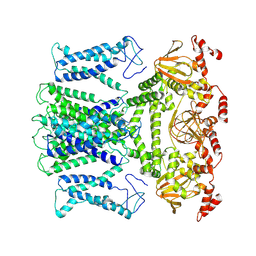 | | Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana | | Descriptor: | POTASSIUM ION, Potassium channel AKT1, Potassium channel KAT3 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Yang, F, Zhang, Y.M, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7VR1
 
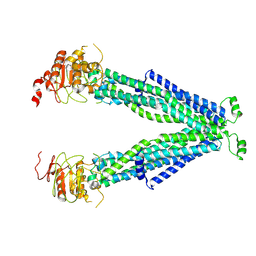 | |
6LQG
 
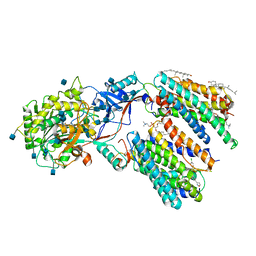 | | Human gamma-secretase in complex with small molecule Avagacestat | | Descriptor: | (2R)-2-[(4-chlorophenyl)sulfonyl-[[2-fluoranyl-4-(1,2,4-oxadiazol-3-yl)phenyl]methyl]amino]-5,5,5-tris(fluoranyl)pentanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7WJ8
 
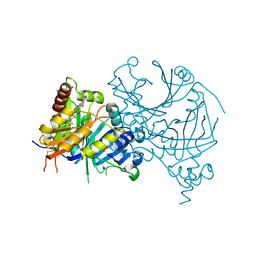 | | Complex structure of AtHPPD-PyQ1 | | Descriptor: | 2-pyren-1-yloxyethyl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-01-05 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7WJJ
 
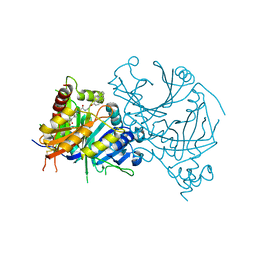 | | Complex structure of AtHPPD-PyQ2 | | Descriptor: | 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]-N-(2-pyren-1-yloxyethyl)ethanamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-01-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
8IOY
 
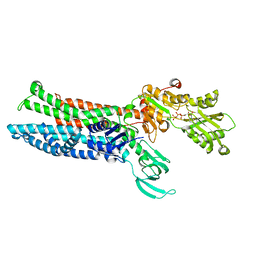 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | Descriptor: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yang, G, Xu, L, Guo, J, Wu, Z. | | Deposit date: | 2023-03-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
