6O9L
 
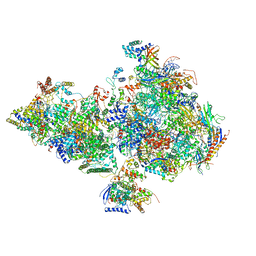 | | Human holo-PIC in the closed state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6O9M
 
 | | Structure of the human apo TFIIH | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription factor IIH subunit 1, General transcription factor IIH subunit 2, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3JB9
 
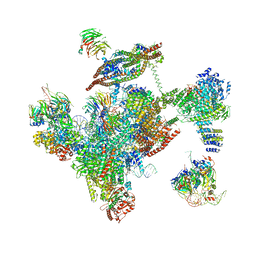 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
5WSG
 
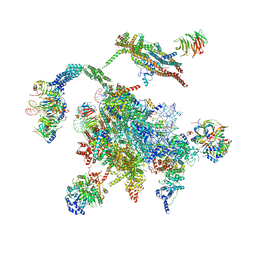 | | Cryo-EM structure of the Catalytic Step II spliceosome (C* complex) at 4.0 angstrom resolution | | Descriptor: | 3'-exon-intron, 3'-intron-lariat, 5'-exon, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a yeast step II catalytically activated spliceosome
Science, 355, 2017
|
|
5GM6
 
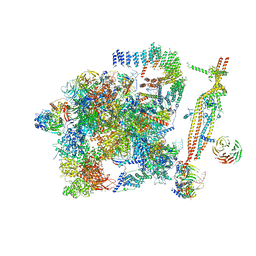 | | Cryo-EM structure of the activated spliceosome (Bact complex) at 3.5 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cold sensitive U2 snRNA suppressor 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a yeast activated spliceosome at 3.5 angstrom resolution
Science, 353, 2016
|
|
7E27
 
 | | Structure of PfFNT in complex with MMV007839 | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | Deposit date: | 2021-02-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
7E26
 
 | | Structure of PfFNT in apo state | | Descriptor: | Formate-nitrite transporter | | Authors: | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | Deposit date: | 2021-02-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
5CQG
 
 | | Structure of Tribolium telomerase in complex with the highly specific inhibitor BIBR1532 | | Descriptor: | 2-{[(2E)-3-(naphthalen-2-yl)but-2-enoyl]amino}benzoic acid, Telomerase reverse transcriptase | | Authors: | Bryan, C, Rice, C, Hoffman, H, Harkisheimer, M, Sweeney, M, Skordalakes, E. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Telomerase Inhibition by the Highly Specific BIBR1532.
Structure, 23, 2015
|
|
6IHU
 
 | |
6IHV
 
 | |
6IHR
 
 | |
6IHS
 
 | |
8IT9
 
 | | Co-crystal structure of FTO bound to 22 | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[(2,6-diethyl-4-pyridin-4-yl-phenyl)amino]-6-(1,4-oxazepan-4-ylmethyl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2023-03-22 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rational Design of RNA Demethylase FTO Inhibitors with Enhanced Antileukemia Drug-Like Properties.
J.Med.Chem., 66, 2023
|
|
7AYE
 
 | | Crystal structure of the computationally designed chemically disruptable heterodimer LD6-MDM2 | | Descriptor: | Isoform 11 of E3 ubiquitin-protein ligase Mdm2, Thiol:disulfide interchange protein DsbD | | Authors: | Yang, C, Lau, K, Pojer, F, Correia, B.E. | | Deposit date: | 2020-11-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A rational blueprint for the design of chemically-controlled protein switches.
Nat Commun, 12, 2021
|
|
1TFZ
 
 | | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-05-27 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
8WOJ
 
 | |
6IHT
 
 | |
6IHL
 
 | | Crystal structure of bacterial serine phosphatase | | Descriptor: | MAGNESIUM ION, Phosphorylated protein phosphatase | | Authors: | Yang, C.-G, yang, T. | | Deposit date: | 2018-09-30 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Insight into the Mechanism of Staphylococcus aureus Stp1 Phosphatase.
Acs Infect Dis., 5, 2019
|
|
6IHW
 
 | |
6YWC
 
 | | De novo designed protein 4E1H_95 in complex with 101F antibody | | Descriptor: | Antibody 101F, Heavy Chain, light chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
3BU0
 
 | | crystal structure of human ABH2 cross-linked to dsDNA with cofactors | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BTX
 
 | | X-ray structure of human ABH2 bound to dsDNA through active site cross-linking | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*TP*GP*CP*G)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DAP*DTP*DAP*DCP*DA)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BTY
 
 | | Crystal structure of human ABH2 bound to dsDNA containing 1meA through cross-linking away from active site | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DGP*DTP*DTP*DAP*DTP*DAP*DCP*DA)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BTZ
 
 | | Crystal structure of human ABH2 cross-linked to dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*AP*GP*GP*TP*GP*AP*(2YR)P*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DTP*DTP*DAP*DTP*DCP*DAP*DCP*DC)-3') | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BUC
 
 | | X-ray structure of human ABH2 bound to dsDNA with Mn(II) and 2KG | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
