4E55
 
 | |
4E57
 
 | |
4E56
 
 | |
6XFP
 
 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | Descriptor: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
6XLO
 
 | | Crystal structure of bRaf in complex with inhibitor | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-[2-fluoro-4-methyl-5-(7-methyl-8-oxo-7,8-dihydropyrido[2,3-d]pyridazin-3-yl)phenyl]benzamide, IODIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Eigenbrot, C, Wang, W. | | Deposit date: | 2020-06-28 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Targeting KRAS Mutant Cancers via Combination Treatment: Discovery of a 5-Fluoro-4-(3 H )-quinazolinone Aryl Urea pan-RAF Kinase Inhibitor.
J.Med.Chem., 64, 2021
|
|
5K4J
 
 | | Crystal Structure of CDK2 in complex with compound 22 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Cyclin-dependent kinase 2 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
5K4I
 
 | | Crystal Structure of ERK2 in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
7RCO
 
 | |
4XJ0
 
 | | Crystal structure of ERK2 in complex with an inhibitor 14K | | Descriptor: | 1-[(1S)-1-(4-chloro-3-fluorophenyl)-2-hydroxyethyl]-4-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]pyridin-2(1H)-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2015-01-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of highly potent, selective, and efficacious small molecule inhibitors of ERK1/2.
J.Med.Chem., 58, 2015
|
|
7U5B
 
 | | Structure of Human KLK5 bound to anti-KLK5 Fab | | Descriptor: | Kallikrein-5, SULFATE ION, anti-KLK5 Fab Heavy Chain, ... | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Dual antibody inhibition of KLK5 and KLK7 for Netherton syndrome and atopic dermatitis.
Sci Transl Med, 14, 2022
|
|
1FL6
 
 | | THE HAPTEN COMPLEXED GERMLINE PRECURSOR TO SULFIDE OXIDASE CATALYTIC ANTIBODY 28B4 | | Descriptor: | 1-[N-4'-NITROBENZYL-N-4'-CARBOXYBUTYLAMINO]METHYLPHOSPHONIC ACID, ANTIBODY GERMLINE PRECURSOR TO 28B4 | | Authors: | Yin, J, Mundorff, E.C, Yang, P.L, Wendt, K.U, Hanway, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2000-08-11 | | Release date: | 2001-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A comparative analysis of the immunological evolution of antibody 28B4.
Biochemistry, 40, 2001
|
|
1FL5
 
 | | THE UNLIGANDED GERMLINE PRECURSOR TO THE SULFIDE OXIDASE CATALYTIC ANTIBODY 28B4. | | Descriptor: | ANTIBODY GERMLINE PRECURSOR TO ANTIBODY 28B4, SULFATE ION | | Authors: | Yin, J, Mundorff, E.C, Yang, P.L, Wendt, K.U, Hanway, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2000-08-11 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A comparative analysis of the immunological evolution of antibody 28B4.
Biochemistry, 40, 2001
|
|
4S0V
 
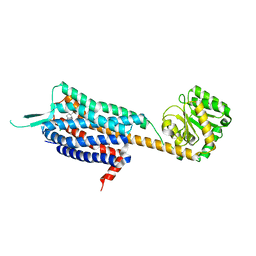 | | Crystal structure of the human OX2 orexin receptor bound to the insomnia drug Suvorexant | | Descriptor: | Human Orexin receptor type 2 fusion protein to P. abysii Glycogen Synthase, OLEIC ACID, [(7R)-4-(5-chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone | | Authors: | Yin, J, Kolb, P, Mobarec, J.C, Rosenbaum, D.M. | | Deposit date: | 2015-01-06 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant.
Nature, 519, 2015
|
|
2Z3D
 
 | |
2Z3E
 
 | |
2Z3C
 
 | |
6VMS
 
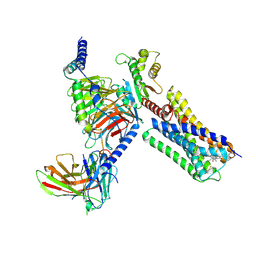 | | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane | | Descriptor: | Endolysin,D(2) dopamine receptor,D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yin, J, Chen, K.M, Clark, M.J, Hijazi, M, Kumari, P, Bai, X, Sunahara, R.K, Barth, P, Rosenbaum, D.M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane.
Nature, 584, 2020
|
|
4ZJ8
 
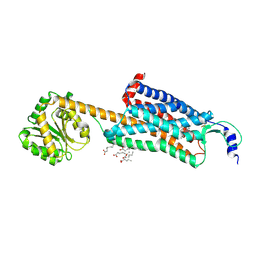 | | Structures of the human OX1 orexin receptor bound to selective and dual antagonists | | Descriptor: | OLEIC ACID, [(7R)-4-(5-chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone, human OX1R fusion protein to P.abysii glycogen synthase | | Authors: | Yin, J, Brautigam, C.A, Shao, Z, Clark, L, Harrell, C.M, Gotter, A.L, Coleman, P, Renger, J.J, Rosenbaum, D.M. | | Deposit date: | 2015-04-29 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6D5Y
 
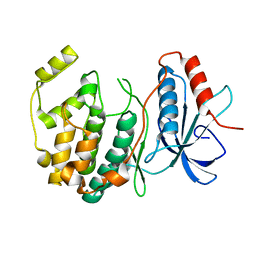 | | Crystal structure of ERK2 G169D mutant | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Jaiswal, B.S, Wang, W. | | Deposit date: | 2018-04-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ERK Mutations and Amplification Confer Resistance to ERK-Inhibitor Therapy.
Clin. Cancer Res., 24, 2018
|
|
4ZJC
 
 | | Structures of the human OX1 orexin receptor bound to selective and dual antagonists | | Descriptor: | OLEIC ACID, [5-(2-fluorophenyl)-2-methyl-1,3-thiazol-4-yl]{(2S)-2-[(5-phenyl-1,3,4-oxadiazol-2-yl)methyl]pyrrolidin-1-yl}methanone, human OX1R fusion protein to P.abysii glycogen synthase | | Authors: | Yin, J, Brautigam, C.A, Shao, Z, Clark, L, Harrell, C.M, Gotter, A.L, Coleman, P.J, Renger, J.J, Rosenbaum, D.M. | | Deposit date: | 2015-04-29 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.832 Å) | | Cite: | Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2HAL
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A Protease 3C, N-ACETYL-LEUCYL-PHENYLALANYL-PHENYLALANYL-GLUTAMATE-FLUOROMETHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
6E59
 
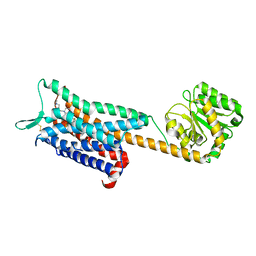 | | Crystal structure of the human NK1 tachykinin receptor | | Descriptor: | 1-(4-{[(2R,3S)-2-{(1R)-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy}-3-(4-fluorophenyl)morpholin-4-yl]methyl}-1H-1,2,3-triazol-5-yl)-N,N-dimethylmethanamine, Substance-P receptor, GlgA glycogen synthase, ... | | Authors: | Yin, J, Clark, L, Chapman, K, Shao, Z, Borek, D, Xu, Q, Wang, J, Rosenbaum, D.M. | | Deposit date: | 2018-07-19 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human NK1tachykinin receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
2H9H
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A virus protease 3C, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Three residue peptide | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-09 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
2CXV
 
 | | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-Derived betaLactone: Selective Crystallization and High-resolution Structure of the His-102 Adduct | | Descriptor: | N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Probable protein P3C | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-07-01 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
