6JJ5
 
 | | BRD4 in complex with 259 | | 分子名称: | 3-((1-methyl-2-oxo-1,2,2a1,5a-tetrahydro-6H-pyrido[3',2':6,7]azepino[4,3,2-cd]isoindol-6-yl)methyl)benzamide, Bromodomain-containing protein 4 | | 著者 | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | 登録日 | 2019-02-25 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | BRD4 in complex with ZZM1
To Be Published
|
|
6JCA
 
 | |
6JC8
 
 | |
6KJ6
 
 | |
7DRX
 
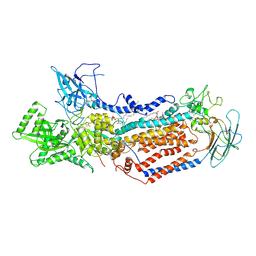 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with beryllium fluoride (E2P state) | | 分子名称: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Xu, J, He, Y, Wu, X, Li, L. | | 登録日 | 2020-12-30 | | 公開日 | 2022-03-23 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSH
 
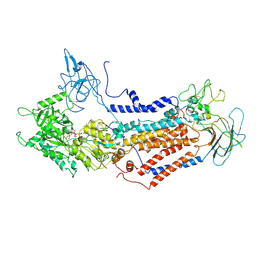 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with AMPPCP (E1-ATP state) | | 分子名称: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | 著者 | Xu, J, He, Y, Wu, X, Li, L. | | 登録日 | 2020-12-31 | | 公開日 | 2022-03-23 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSI
 
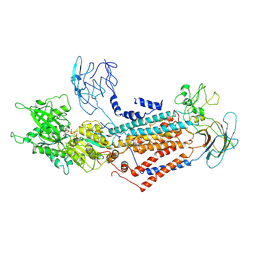 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with AMPPCP ( resting state ) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | 著者 | Xu, J, He, Y, Wu, X, Li, L. | | 登録日 | 2020-12-31 | | 公開日 | 2022-03-23 | | 最終更新日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7EJK
 
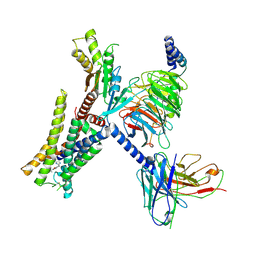 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to oxymetazoline | | 分子名称: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, J, Cao, S, Liu, Z, Du, Y. | | 登録日 | 2021-04-02 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJ0
 
 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex | | 分子名称: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, J, Cao, S, Liu, Z, Du, Y. | | 登録日 | 2021-04-01 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJA
 
 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to dexmedetomidine | | 分子名称: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Xu, J, Cao, S, Liu, Z, Du, Y. | | 登録日 | 2021-04-01 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJ8
 
 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to brimonidine | | 分子名称: | Alpha-2A adrenergic receptor, Brimonidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Xu, J, Cao, S, Liu, Z, Du, Y. | | 登録日 | 2021-04-01 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJX
 
 | | Structure of the GPR88-Gi1 signaling complex bound to a synthetic ligand | | 分子名称: | (1R,2R)-N-[(2S,3S)-2-azanyl-3-methyl-pentyl]-N-[4-(4-propylphenyl)phenyl]-2-pyridin-2-yl-cyclopropane-1-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Xu, J, Chen, G, Liu, Z, Du, Y. | | 登録日 | 2021-04-02 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Activation and allosteric regulation of the orphan GPR88-Gi1 signaling complex.
Nat Commun, 13, 2022
|
|
6ISK
 
 | | SnoaL-like cyclase XimE | | 分子名称: | 1,2-ETHANEDIOL, MALONIC ACID, XimE, ... | | 著者 | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | 登録日 | 2018-11-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
6ISL
 
 | | SnoaL-like cyclase XimE with its product xiamenmycin B | | 分子名称: | (2R,3S)-2-methyl-2-(4-methylpent-3-enyl)-3-oxidanyl-3,4-dihydrochromene-6-carboxylic acid, XimE, SnoaL-like domain protein | | 著者 | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | 登録日 | 2018-11-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
197L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | 登録日 | 1995-11-06 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
6PWW
 
 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | 分子名称: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | 登録日 | 2019-07-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
2A8H
 
 | | Crystal structure of catalytic domain of TACE with Thiomorpholine Sulfonamide Hydroxamate inhibitor | | 分子名称: | 4-({4-[(4-AMINOBUT-2-YNYL)OXY]PHENYL}SULFONYL)-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | 著者 | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Jin, G, Barone, D, Skotnicki, J.S. | | 登録日 | 2005-07-08 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Acetylenic TACE inhibitors. Part 3: Thiomorpholine sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ZXC
 
 | | Crystal structure of catalytic domain of TNF-alpha converting enzyme (TACE) with inhibitor | | 分子名称: | (3S)-4-{[4-(BUT-2-YNYLOXY)PHENYL]SULFONYL}-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | 著者 | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Zhang, Y, Jin, G, Cowling, R, Barone, D, Skotnicki, J.S. | | 登録日 | 2005-06-07 | | 公開日 | 2005-09-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Acetylenic TACE inhibitors. Part 2: SAR of six-membered cyclic sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | 分子名称: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | 登録日 | 2019-07-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | 登録日 | 2020-07-15 | | 公開日 | 2020-09-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6ZYA
 
 | | Extended human uromodulin filament core at 3.5 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | 登録日 | 2020-07-31 | | 公開日 | 2020-09-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6PWX
 
 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | 分子名称: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | 登録日 | 2019-07-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6H67
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | 登録日 | 2018-07-26 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6H68
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49 | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | 登録日 | 2018-07-26 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | 分子名称: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | 著者 | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
