4XKL
 
 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | Descriptor: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
5WDY
 
 | |
5SUN
 
 | | IDH1 R132H in complex with IDH146 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-N-[3-(dimethylsulfamoyl)phenyl]-4-oxo-3,4-dihydrophthalazine-1-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.477 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5SVF
 
 | | IDH1 R132H in complex with IDH125 | | Descriptor: | (4S)-3-(2-{[(1S)-1-phenylethyl]amino}pyrimidin-4-yl)-4-(propan-2-yl)-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5SVO
 
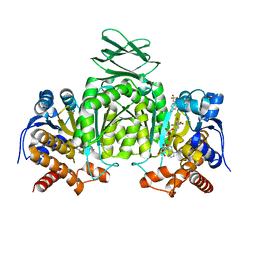 | | Structure of IDH2 mutant R140Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-06 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5SVN
 
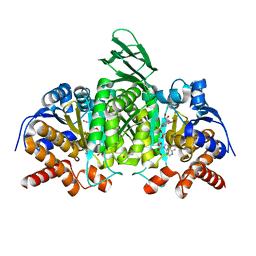 | | Structure of IDH2 mutant R172K | | Descriptor: | DI(HYDROXYETHYL)ETHER, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-06 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
1JUO
 
 | |
1JNK
 
 | |
1PME
 
 | |
1TAF
 
 | | DROSOPHILA TBP ASSOCIATED FACTORS DTAFII42/DTAFII62 HETEROTETRAMER | | Descriptor: | TFIID TBP ASSOCIATED FACTOR 42, TFIID TBP ASSOCIATED FACTOR 62, ZINC ION | | Authors: | Xie, X, Kokubo, T, Cohen, S.L, Mirza, U.A, Hoffmann, A, Chait, B.T, Roeder, R.G, Nakatani, Y, Burley, S.K. | | Deposit date: | 1996-06-01 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural similarity between TAFs and the heterotetrameric core of the histone octamer.
Nature, 380, 1996
|
|
1TKQ
 
 | | SOLUTION STRUCTURE OF A LINKED UNSYMMETRIC GRAMICIDIN IN A MEMBRANE-ISOELECTRICAL SOLVENTS MIXTURE IN THE PRESENCE OF CsCl | | Descriptor: | GRAMICIDIN A, MINI-GRAMICIDIN A, SUCCINIC ACID | | Authors: | Xie, X, Al-Momani, L, Bockelmann, D, Griesinger, C, Koert, U. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-13 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | An Asymmetric Ion Channel Derived from Gramicidin A. Synthesis, Function and NMR Structure.
FEBS J., 272, 2005
|
|
5TF9
 
 | |
6B0Z
 
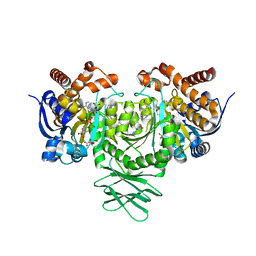 | | IDH1 R132H mutant in complex with IDH305 | | Descriptor: | (4R)-4-[(1S)-1-fluoroethyl]-3-[2-({(1S)-1-[4-methyl-2'-(trifluoromethyl)[3,4'-bipyridin]-6-yl]ethyl}amino)pyrimidin-4-yl]-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2017-09-15 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Discovery and Evaluation of Clinical Candidate IDH305, a Brain Penetrant Mutant IDH1 Inhibitor.
ACS Med Chem Lett, 8, 2017
|
|
3G90
 
 | |
5TQH
 
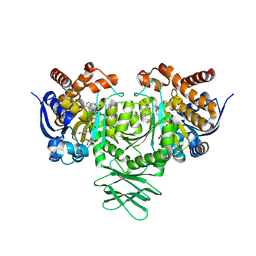 | | IDH1 R132H mutant in complex with IDH889 | | Descriptor: | (4S)-3-[2-({(1S)-1-[5-(4-fluoro-3-methylphenyl)pyrimidin-2-yl]ethyl}amino)pyrimidin-4-yl]-4-(propan-2-yl)-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 3-Pyrimidin-4-yl-oxazolidin-2-ones as Allosteric and Mutant Specific Inhibitors of IDH1.
ACS Med Chem Lett, 8, 2017
|
|
5WE8
 
 | |
2OJG
 
 | |
2OJI
 
 | |
2OK1
 
 | |
2OJJ
 
 | |
6KR4
 
 | | Crystal structure of the liprin-alpha3_SAM123/LAR_D1D2 complex | | Descriptor: | HEXAETHYLENE GLYCOL, Liprin-alpha-3, PHOSPHATE ION, ... | | Authors: | Xie, X, Liang, M, Luo, L, Wei, Z. | | Deposit date: | 2019-08-20 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of liprin-alpha-promoted LAR-RPTP clustering for modulation of phosphatase activity.
Nat Commun, 11, 2020
|
|
4IEH
 
 | |
3B70
 
 | | Crystal structure of Aspergillus terreus trans-acting lovastatin polyketide enoyl reductase (LovC) with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6EG3
 
 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6EG2
 
 | | Crystal structure of human BRM in complex with compound 16 | | Descriptor: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
