8HFF
 
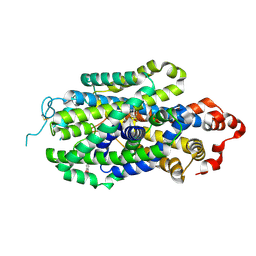 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of norepinephrine in an inward-open state at resolution of 2.9 angstrom. | | Descriptor: | L-NOREPINEPHRINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Molecular basis for the reuptake and inhibition of human norepinephrine transporter
To Be Published
|
|
8HFL
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant bupropion in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | Bupropion, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-11 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the reuptake and inhibition of human norepinephrine transporter
To Be Published
|
|
8HFI
 
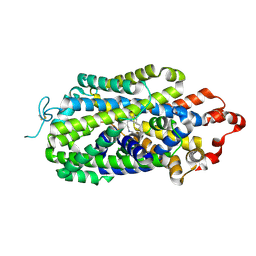 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant desipramine in an inward-open state at resolution of 2.5 angstrom. | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis for the reuptake and inhibition of human norepinephrine transporter
To Be Published
|
|
8HFE
 
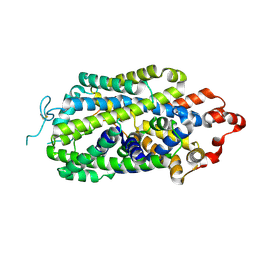 | | Cryo-EM structure of human norepinephrine transporter NET in an inward-open state at resolution of 2.5 angstrom | | Descriptor: | CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis for the reuptake and inhibition of human norepinephrine transporter
To Be Published
|
|
8HFG
 
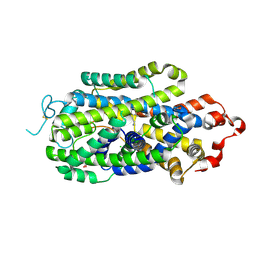 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of dopamine in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | CHLORIDE ION, L-DOPAMINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the reuptake and inhibition of human norepinephrine transporter
To Be Published
|
|
5H9F
 
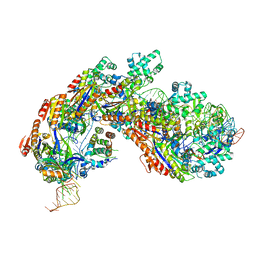 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target at 2.45 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
5FB5
 
 | |
5FEI
 
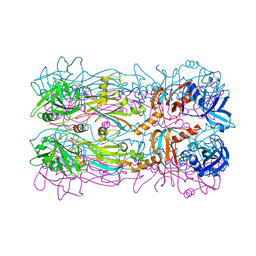 | | Crystal structure of the bacteriophage phi29 tail knob protein gp9 truncation variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Distal tube protein | | Authors: | Xu, J.W, Gui, M, Wang, D.H, Xiang, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The bacteriophage 29 tail possesses a pore-forming loop for cell membrane penetration.
Nature, 534, 2016
|
|
5FB4
 
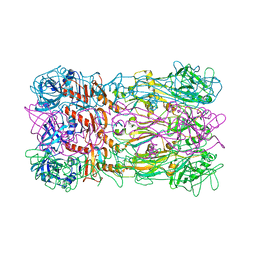 | | Crystal structure of the bacteriophage phi29 tail knob protein gp9 truncation variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Distal tube protein | | Authors: | Xu, J.W, Gui, M, Wang, D.H, Xiang, Y. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | The bacteriophage 29 tail possesses a pore-forming loop for cell membrane penetration.
Nature, 534, 2016
|
|
6LUM
 
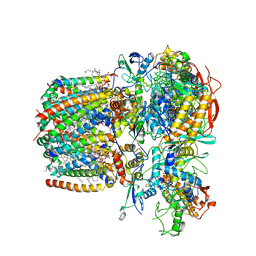 | | Structure of Mycobacterium smegmatis succinate dehydrogenase 2 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Gao, Y, Gong, H, Zhou, X, Xiao, Y, Wang, W, Ji, W, Wang, Q, Rao, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Nat Commun, 11, 2020
|
|
5H9E
 
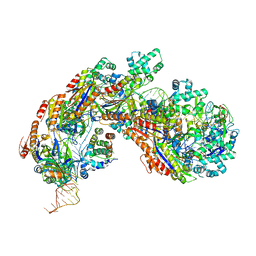 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target (32-nt spacer) at 3.20 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
4ZM6
 
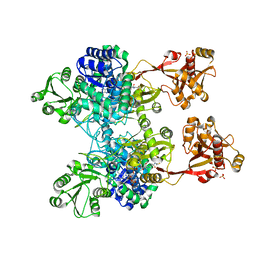 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
3O5Z
 
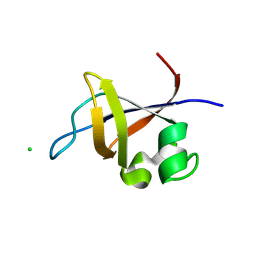 | | Crystal structure of the SH3 domain from p85beta subunit of phosphoinositide 3-kinase (PI3K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Chen, S, Xiao, Y, Ponnusamy, R, Tan, J, Lei, J, Hilgenfeld, R. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-ray structure of the SH3 domain of the phosphoinositide 3-kinase p85 beta subunit
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5YJ7
 
 | | Structural insight into the beta-GH1 glucosidase BGLN1 from oleaginous microalgae Nannochloropsis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside hydrolase | | Authors: | Dong, S, Liu, Y.J, Zhou, H.X, Xiao, Y, Xu, J, Cui, Q, Wang, X.Q, Feng, Y.G. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insight into a GH1 beta-glucosidase from the oleaginous microalga, Nannochloropsis oceanica.
Int.J.Biol.Macromol., 170, 2021
|
|
4KFN
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
5ZMD
 
 | | Crystal structure of FTO in complex with m6dA modified ssDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, DNA (5'-D(P*TP*CP*TP*(6MA)P*TP*AP*TP*CP*G)-3'), MANGANESE (II) ION, ... | | Authors: | Zhang, X, Wei, L.H, Luo, J, Xiao, Y, Liu, J, Zhang, W, Zhang, L, Jia, G.F. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4KFO
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Tosteb, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
1U9S
 
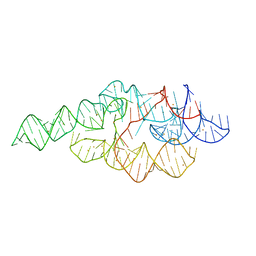 | | Crystal structure of the specificity domain of Ribonuclease P of the A-type | | Descriptor: | BARIUM ION, RIBONUCLEASE P | | Authors: | Krasilnikov, A.S, Xiao, Y, Pan, T, Mondragon, A. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Basis for structural diversity in homologous RNAs.
Science, 306, 2004
|
|
7K3A
 
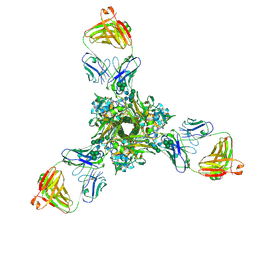 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation B, fusion peptide release | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K3B
 
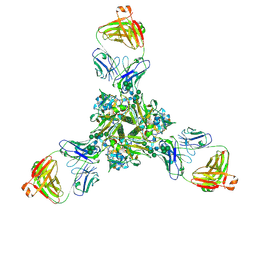 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation C, central helices splay | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K37
 
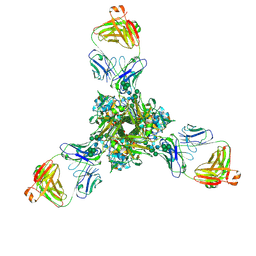 | | Structure of full-length influenza HA with a head-binding antibody at pH 7.8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Xiang, Y, Gao, J. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K39
 
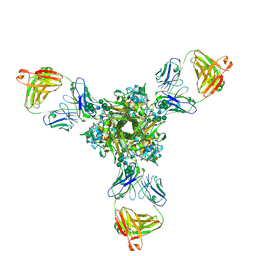 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation A, neutral pH-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
1X7I
 
 | | Crystal structure of the native copper homeostasis protein (cutCm) with calcium binding from Shigella flexneri 2a str. 301 | | Descriptor: | CALCIUM ION, Copper homeostasis protein cutC | | Authors: | Zhu, D.Y, Zhu, Y.Q, Huang, R.H, Xiang, Y, Wang, D.C. | | Deposit date: | 2004-08-14 | | Release date: | 2005-03-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the copper homeostasis protein (CutCm) from Shigella flexneri at 1.7 A resolution: The first structure of a new sequence family of TIM barrels
Proteins, 58, 2004
|
|
4EKX
 
 | |
5WRG
 
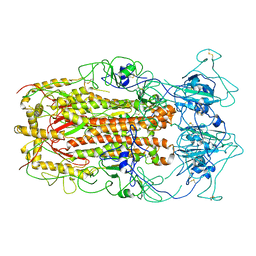 | | SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-11 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
