1EGO
 
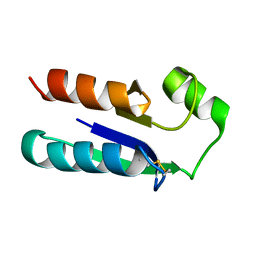 | | NMR STRUCTURE OF OXIDIZED ESCHERICHIA COLI GLUTAREDOXIN: COMPARISON WITH REDUCED E. COLI GLUTAREDOXIN AND FUNCTIONALLY RELATED PROTEINS | | Descriptor: | GLUTAREDOXIN | | Authors: | Xia, T.-H, Bushweller, J.H, Sodano, P, Billeter, M, Bjornberg, O, Holmgren, A, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
5WUZ
 
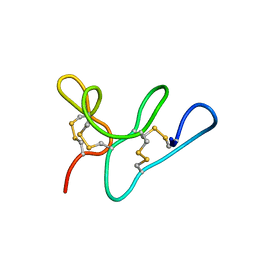 | | Morintides mO1 | | Descriptor: | Morintide mO1 | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Morintides: 8C-Hevein-like peptides without a protein cargo from the drumstick tree Moringa oleifera
To Be Published
|
|
1QWO
 
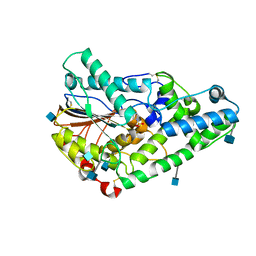 | | Crystal structure of a phosphorylated phytase from Aspergillus fumigatus, revealing the structural basis for its heat resilience and catalytic pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Xiang, T, Liu, Q, Deacon, A.M, Koshy, M, Kriksunov, I.A, Lei, X.G, Hao, Q, Thiel, D.J. | | Deposit date: | 2003-09-03 | | Release date: | 2004-06-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Heat-resilient Phytase from Aspergillus fumigatus, Carrying a Phosphorylated Histidine
J.Mol.Biol., 339, 2004
|
|
6BZ9
 
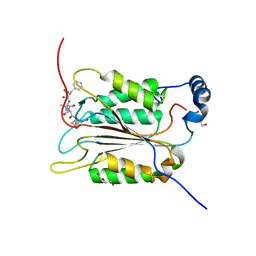 | | Crystal structure of human caspase-1 in complex with Ac-FLTD-CMK | | Descriptor: | Ac-FLTD-CMK, Caspase-1, DI(HYDROXYETHYL)ETHER | | Authors: | Xiao, T.S, Yang, J, Liu, Z, Wang, C, Yang, R. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6V4T
 
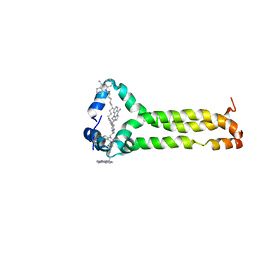 | | MPER-TMD of HIV-1 Env bound with the entry inhibitor S2C3 | | Descriptor: | 4,4'-(decane-1,10-diyl)bis(9-amino-2,3-dihydro-1H-cyclopenta[b]quinolin-4-ium), Envelope glycoprotein gp160 | | Authors: | Xiao, T, Frey, G, Fu, Q, Lavine, C.L, Scott, D.A, Seaman, M.S, Chou, J.J, Chen, B. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes.
Nat.Chem.Biol., 16, 2020
|
|
1D2Z
 
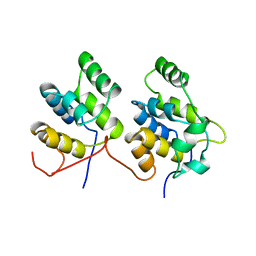 | | THREE-DIMENSIONAL STRUCTURE OF A COMPLEX BETWEEN THE DEATH DOMAINS OF PELLE AND TUBE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DEATH DOMAIN OF PELLE, DEATH DOMAIN OF TUBE | | Authors: | Xiao, T, Towb, P, Wasserman, S.A, Sprang, S.R. | | Deposit date: | 1999-09-28 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a complex between the death domains of Pelle and Tube.
Cell(Cambridge,Mass.), 99, 1999
|
|
5XBD
 
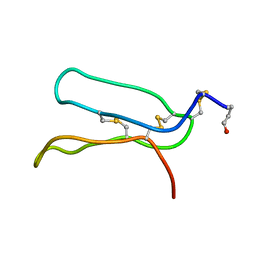 | |
5XDI
 
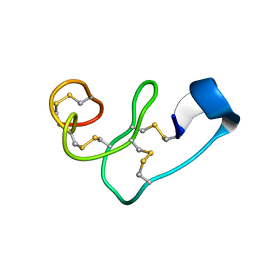 | |
5GSF
 
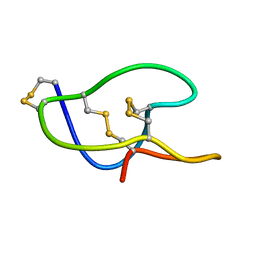 | | Structure of roseltide rT1 | | Descriptor: | roseltide rT1 | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of Roseltide, a Knottin-type Neutrophil Elastase Inhibitor Derived from Hibiscus sabdariffa.
Sci Rep, 6, 2016
|
|
2FTP
 
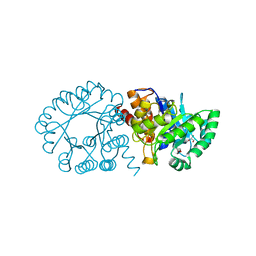 | | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SODIUM ION, hydroxymethylglutaryl-CoA lyase | | Authors: | Xiao, T, Evdokimova, E, Liu, Y, Kudritska, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-24 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa
To be Published
|
|
1TYE
 
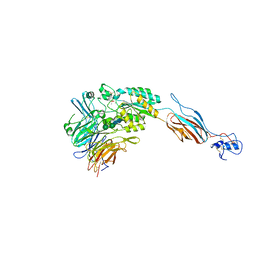 | | Structural basis for allostery in integrins and binding of ligand-mimetic therapeutics to the platelet receptor for fibrinogen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Xiao, T, Takagi, J, Coller, B.S, Wang, J.-H, Springer, T.A. | | Deposit date: | 2004-07-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics
Nature, 432, 2004
|
|
1IK7
 
 | | Crystal Structure of the Uncomplexed Pelle Death Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PROBABLE SERINE/THREONINE-PROTEIN KINASE Pelle | | Authors: | Xiao, T, Gardner, K.H, Sprang, S.R. | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cosolvent-induced transformation
of a death domain tertiary structure
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5H1H
 
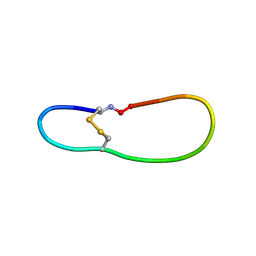 | | NMR structure of SLBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
5H1I
 
 | | NMR structure of TIBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
1EDY
 
 | |
7F24
 
 | |
7F0T
 
 | |
7F23
 
 | |
7F1O
 
 | |
7F1Z
 
 | |
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
