4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
3O5X
 
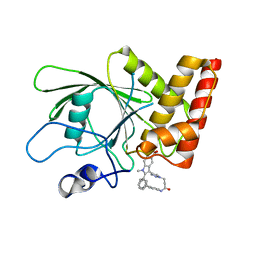 | | Crystal structure of the oncogenic tyrosine phosphatase SHP2 complexed with a salicylic acid-based small molecule inhibitor | | Descriptor: | 3-{1-[3-(biphenyl-4-ylamino)-3-oxopropyl]-1H-1,2,3-triazol-4-yl}-6-hydroxy-1-methyl-2-phenyl-1H-indole-5-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Zhang, X, He, Y, Liu, S, Yu, Z, Jiang, Z, Yang, Z, Dong, Y, Nabinger, S.C, Wu, L, Gunawan, A.M, Wang, L, Chan, R.J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).
J.Med.Chem., 53, 2010
|
|
7PSI
 
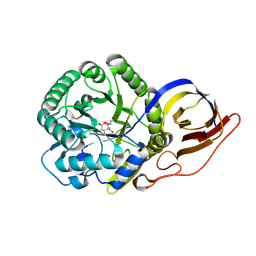 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor ME727 | | Descriptor: | (2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase, SULFATE ION | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PSK
 
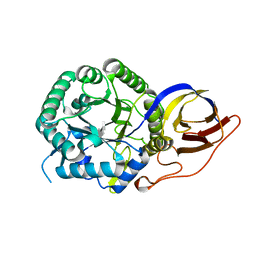 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor GR109 | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PSJ
 
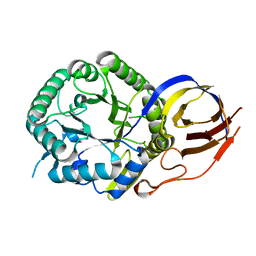 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VL166 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase | | Authors: | Armstrong, Z, Wu, L, Davies, G.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PSH
 
 | |
7PLS
 
 | | Cryo-EM structures of human fucosidase FucA1 reveal insight into substate recognition and catalysis. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tissue alpha-L-fucosidase | | Authors: | Armstrong, Z, Meek, R.W, Wu, L, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-09-01 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structures of human fucosidase FucA1 reveal insight into substrate recognition and catalysis.
Structure, 30, 2022
|
|
7PM4
 
 | | Cryo-EM structures of human fucosidase FucA1 reveal insight into substate recognition and catalysis. | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Tissue alpha-L-fucosidase | | Authors: | Armstrong, Z, Meek, R.W, Wu, L, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-09-01 | | Release date: | 2022-08-10 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structures of human fucosidase FucA1 reveal insight into substrate recognition and catalysis.
Structure, 30, 2022
|
|
8WKY
 
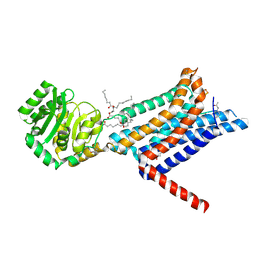 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
8WKZ
 
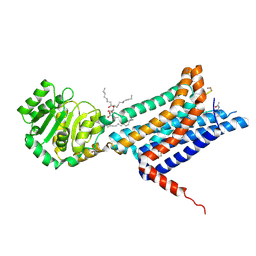 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S31 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
5U3J
 
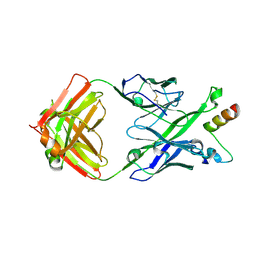 | | Crystal Structure of DH511.1 Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.1 Heavy Chain, DH511.1 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3O
 
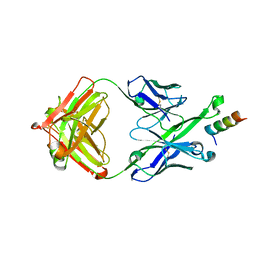 | | Crystal Structure of DH511.2_K3 Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.2_K3 Fab Heavy Chain, DH511.2_K3 Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3K
 
 | | Crystal Structure of DH511.2 Fab in Complex with HIV-1 gp41 MPER 662-683 Peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, DH511.2 Fab Heavy Chain, ... | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3N
 
 | | Crystal Structure of DH511.12P Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.12P Fab Heavy Chain, DH511.12P Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3M
 
 | | Crystal Structure of DH511.11P Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.11P Fab Heavy Chain, DH511.11P Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.418 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
4GA7
 
 | | Crystal structure of human serpinB1 mutant | | Descriptor: | Leukocyte elastase inhibitor | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4GAW
 
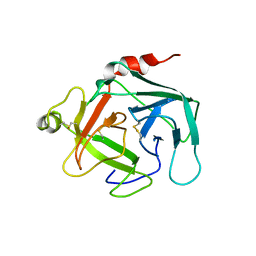 | | Crystal structure of active human granzyme H | | Descriptor: | CHLORIDE ION, Granzyme H, SULFATE ION | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
8OHR
 
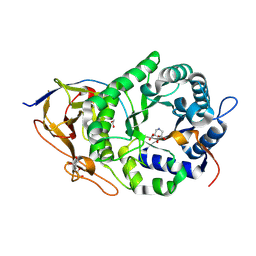 | | Crystal structure of human heparanase in complex with glucuronic acid configured 3-geminal diol iminosugar inhibitor | | Descriptor: | (3~{S},4~{R})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHW
 
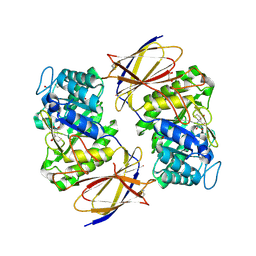 | | Crystal structure of heparanase from Burkholderia pseudomallei in complex with siastatin B derived inhibitor | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, Glycoside hydrolase family 44 domain-containing protein | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
4DGX
 
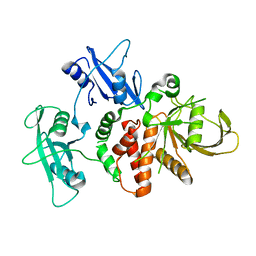 | | LEOPARD Syndrome-Associated SHP2/Y279C mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
8OHV
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured 3-geminal diol iminosugar inhibitor | | Descriptor: | (3~{S},4~{R})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHX
 
 | | Crystal structure of Beta-glucuronidase from Escherichia coli in complex with siastatin B derived inhibitor | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OGX
 
 | | Beta-glucuronidase from Acidobacterium capsulatum in complex with inhibitor R3794 | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, PHOSPHATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHT
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHU
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured isofagamine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
