2NCK
 
 | |
1LMK
 
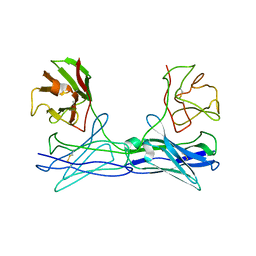 | | THE STRUCTURE OF A BIVALENT DIABODY | | Descriptor: | ANTI-PHOSPHATIDYLINOSITOL SPECIFIC PHOSPHOLIPASE C DIABODY | | Authors: | Williams, R.L. | | Deposit date: | 1994-08-29 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a diabody, a bivalent antibody fragment.
Structure, 2, 1994
|
|
1NLK
 
 | |
1NQB
 
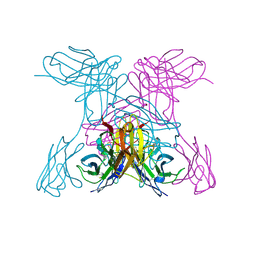 | | TRIVALENT ANTIBODY FRAGMENT | | Descriptor: | SINGLE-CHAIN ANTIBODY FRAGMENT | | Authors: | Williams, R.L, Pei, X.Y. | | Deposit date: | 1997-04-04 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0-A resolution crystal structure of a trimeric antibody fragment with noncognate VH-VL domain pairs shows a rearrangement of VH CDR3.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1NSK
 
 | |
1RBB
 
 | |
5OQ4
 
 | | PQR309 - a Potent, Brain-Penetrant, Orally Bioavailable, pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology | | Descriptor: | 5-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Williams, R.L, Zhang, X. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology.
J. Med. Chem., 60, 2017
|
|
2UZI
 
 | | Crystal structure of HRAS(G12V) - anti-RAS Fv complex | | Descriptor: | ANTI-RAS FV HEAVY CHAIN, ANTI-RAS FV LIGHT CHAIN, GTPASE HRAS, ... | | Authors: | Tanaka, T, williams, R.L, Rabbitts, T.H. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tumour Prevention by a Single Antibody Domain Targeting the Interaction of Signal Transduction Proteins with Ras.
Embo J., 26, 2007
|
|
2V6X
 
 | | Stractural insight into the interaction between ESCRT-III and Vps4 | | Descriptor: | DOA4-INDEPENDENT DEGRADATION PROTEIN 4, SULFATE ION, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4 | | Authors: | Obita, T, Perisic, O, Ghazi-Tabatabai, S, Saksena, S, Emr, S.D, Williams, R.L. | | Deposit date: | 2007-07-23 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis for Selective Recognition of Escrt-III by the Aaa ATPase Vps4
Nature, 449, 2007
|
|
2V1Y
 
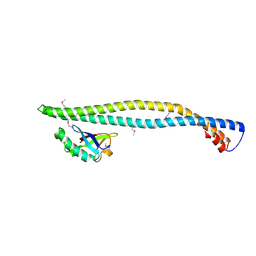 | | Structure of a phosphoinositide 3-kinase alpha adaptor-binding domain (ABD) in a complex with the iSH2 domain from p85 alpha | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Miled, N, Yan, Y, Hon, W.C, Perisic, O, Zvelebil, M, Inbar, Y, Schneidman-Duhovny, D, Wolfson, H.J, Backer, J.M, Williams, R.L. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit.
Science, 317, 2007
|
|
8OW2
 
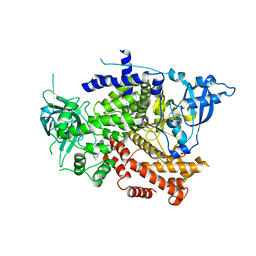 | | Crystal structure of the p110alpha catalytic subunit from homo sapiens in complex with activator 1938 | | Descriptor: | 1-[7-[[2-[[4-(4-ethylpiperazin-1-yl)phenyl]amino]pyridin-4-yl]amino]-2,3-dihydroindol-1-yl]ethanone, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
2CHZ
 
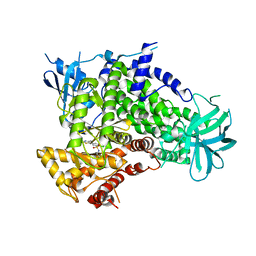 | | A pharmacological map of the PI3-K family defines a role for p110alpha in signaling: The structure of complex of phosphoinositide 3-kinase gamma with inhibitor PIK-93 | | Descriptor: | N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
2CHW
 
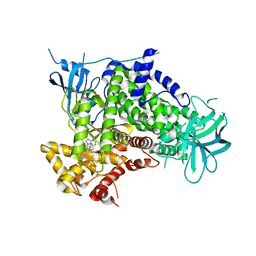 | | A pharmacological map of the PI3-K family defines a role for p110 alpha in signaling: The structure of complex of phosphoinositide 3- kinase gamma with inhibitor PIK-39 | | Descriptor: | 2-((9H-PURIN-6-YLTHIO)METHYL)-5-CHLORO-3-(2-METHOXYPHENYL)QUINAZOLIN-4(3H)-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
2CHX
 
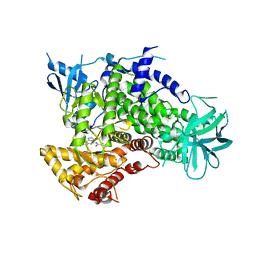 | | A pharmacological map of the PI3-K family defines a role for p110alpha in signaling: The structure of complex of phosphoinositide 3-kinase gamma with inhibitor PIK-90 | | Descriptor: | N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
8RCH
 
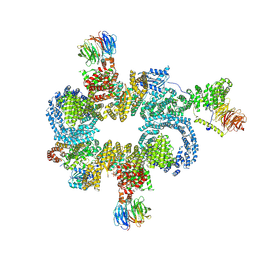 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, overall refinement | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | cryoEM structure of mTORC1 with the cancer-associated 1455-EWED-1458 duplication variant, overall refinement
To Be Published
|
|
8RCK
 
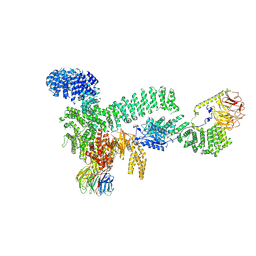 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused on one protomer copy. | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused on one protomer copy.
To Be Published
|
|
8RCN
 
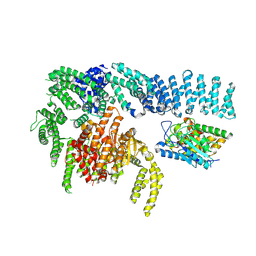 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused region of mTOR and RAPTOR on one protomer copy. | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused region of mTOR and RAPTOR on one protomer copy.
To Be Published
|
|
7BL1
 
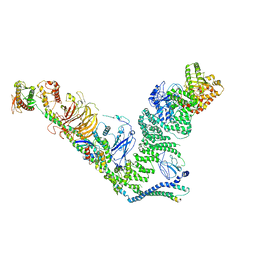 | | human complex II-BATS bound to membrane-attached Rab5a-GTP | | Descriptor: | Beclin-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tremel, S, Morado, D.R, Kovtun, O, Williams, R.L, Briggs, J.A.G, Munro, S, Ohashi, Y, Bertram, J, Perisic, O. | | Deposit date: | 2021-01-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes.
Nat Commun, 12, 2021
|
|
8OXP
 
 | | ATM(Q2971A) in complex with Mg AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine-protein kinase ATM, ... | | Authors: | Howes, A.C, Perisic, O, Williams, R.L. | | Deposit date: | 2023-05-02 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress.
Sci Adv, 9, 2023
|
|
8OXQ
 
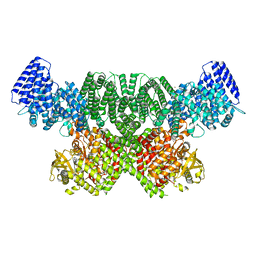 | | ATM(Q2971A) dimeric C-terminal region in complex with Mg AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine-protein kinase ATM, ... | | Authors: | Howes, A.C, Perisic, O, Williams, R.L. | | Deposit date: | 2023-05-02 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress.
Sci Adv, 9, 2023
|
|
8OXM
 
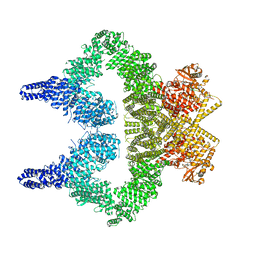 | | ATM(Q2971A) activated by oxidative stress in complex with Mg AMP-PNP and p53 peptide | | Descriptor: | Cellular tumor antigen p53, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Howes, A.C, Perisic, O, Williams, R.L. | | Deposit date: | 2023-05-02 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress.
Sci Adv, 9, 2023
|
|
8OXO
 
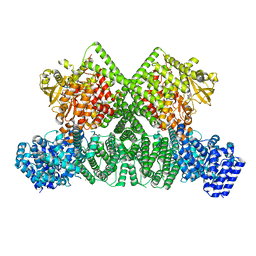 | | ATM(Q2971A) dimeric C-terminal region activated by oxidative stress in complex with Mg AMP-PNP and p53 peptide | | Descriptor: | Cellular tumor antigen p53, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Howes, A.C, Perisic, O, Williams, R.L. | | Deposit date: | 2023-05-02 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress.
Sci Adv, 9, 2023
|
|
6S6D
 
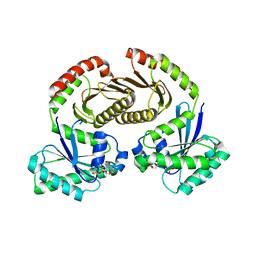 | | Crystal structure of RagA-Q66L-GTP/RagC-S75N-GDP GTPase heterodimer complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Anandapadamanaban, M, Masson, G.R, Perisic, O, Kaufman, J, Williams, R.L. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
6S6A
 
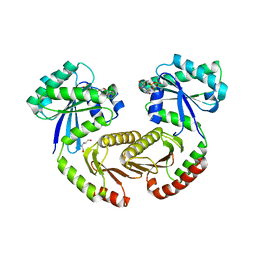 | | Crystal structure of RagA-Q66L/RagC-T90N GTPase heterodimer complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Anandapadamanaban, M, Masson, G.R, Perisic, O, Kaufman, J, Williams, R.L. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
6SB2
 
 | | cryo-EM structure of mTORC1 bound to active RagA/C GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Ras-related GTP-binding protein A, ... | | Authors: | Anandapadamanaban, M, Berndt, A, Masson, G.R, Perisic, O, Williams, R.L. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
