3TIK
 
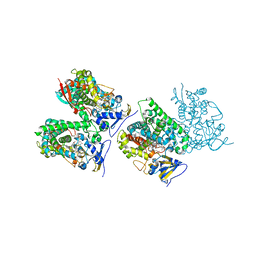 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in complex with the tipifarnib derivative 6-((4-chlorophenyl)(methoxy)(1-methyl-1H-imidazol-5-yl)methyl)-4-(2,6-difluorophenyl)-1-methylquinolin-2(1H)-one | | Descriptor: | 6-[(R)-(4-chlorophenyl)(methoxy)(1-methyl-1H-imidazol-5-yl)methyl]-4-(2,6-difluorophenyl)-1-methylquinolin-2(1H)-one, PROTOPORPHYRIN IX CONTAINING FE, sterol 14-alpha demethylase (CYP51) | | Authors: | Hargrove, T.Y, Wawrzak, Z, Kraus, J.M, Gelb, M.H, Buckner, F.S, Waterman, M.R, Lepesheva, G.I. | | Deposit date: | 2011-08-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pharmacological characterization, structural studies, and in vivo activities of anti-chagas disease lead compounds derived from tipifarnib.
Antimicrob.Agents Chemother., 56, 2012
|
|
5UXC
 
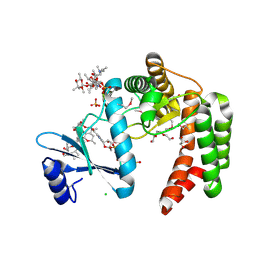 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with GDP | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXD
 
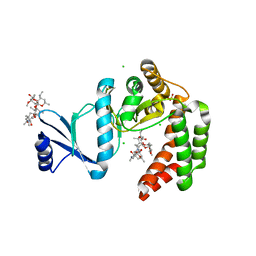 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with azithromycin | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, Macrolide 2'-phosphotransferase MphH, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5W6L
 
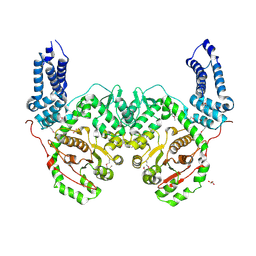 | | Crystal Structure of RRSP, a MARTX Toxin Effector Domain from Vibrio vulnificus CMCP6 | | Descriptor: | CHLORIDE ION, GLYCEROL, RTX repeat-containing cytotoxin, ... | | Authors: | Minasov, G, Wawrzak, Z, Biancucci, M, Shuvalova, L, Dubrovska, I, Satchell, K.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The bacterial Ras/Rap1 site-specific endopeptidase RRSP cleaves Ras through an atypical mechanism to disrupt Ras-ERK signaling.
Sci Signal, 11, 2018
|
|
5WOL
 
 | | Crystal structure of dihydrodipicolinate reductase DapB from Coxiella burnetii | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 4-hydroxy-tetrahydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase DapB from Coxiella burnetii
To Be Published
|
|
4H5F
 
 | | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1
TO BE PUBLISHED
|
|
3OQ7
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OQA
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4DD5
 
 | | Biosynthetic Thiolase (ThlA1) from Clostridium difficile | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biosynthetic Thiolase (ThlA1) from Clostridium difficile
To be Published
|
|
6BVC
 
 | | Crystal structure of AAC(3)-Ia in complex with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside-(3)-N-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | To be published
To Be Published
|
|
3GOA
 
 | | Crystal structure of the Salmonella typhimurium FadA 3-ketoacyl-CoA thiolase | | Descriptor: | 3-ketoacyl-CoA thiolase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Anderson, S.M, Skarina, T, Onopriyenko, O, Wawrzak, Z, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: |
|
|
3IGS
 
 | | Structure of the Salmonella enterica N-acetylmannosamine-6-phosphate 2-epimerase | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, N-acetylmannosamine-6-phosphate 2-epimerase 2, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
5HMN
 
 | | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol | | Descriptor: | AAC3-I, COENZYME A, TETRAETHYLENE GLYCOL | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol
To Be Published
|
|
5VGC
 
 | | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Borek, D, Valleau, D, Skarina, T, Jobin, M.C, Wawrzak, Z, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
6M8T
 
 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidnova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex
To Be Published
|
|
3NX3
 
 | | Crystal structure of acetylornithine aminotransferase (argD) from Campylobacter jejuni | | Descriptor: | Acetylornithine aminotransferase, MAGNESIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-12 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
3Q58
 
 | | Structure of N-acetylmannosamine-6-Phosphate Epimerase from Salmonella enterica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-27 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
5HT0
 
 | | Crystal structure of an Antibiotic_NAT family aminoglycoside acetyltransferase HMB0038 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | Aminoglycoside acetyltransferase HMB0005, COENZYME A, SULFATE ION | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
5F47
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with trehalose | | Descriptor: | CALCIUM ION, CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5TVL
 
 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | Descriptor: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | Authors: | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A
To Be Published
|
|
4F21
 
 | | Crystal structure of carboxylesterase/phospholipase family protein from Francisella tularensis | | Descriptor: | Carboxylesterase/phospholipase family protein, N-((1R,2S)-2-allyl-4-oxocyclobutyl)-4-methylbenzenesulfonamide, bound form | | Authors: | Filippova, E.V, Minasov, G, Kuhn, M, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J.R, Kiryukhina, O, Becker, D.P, Armoush, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large scale structural rearrangement of a serine hydrolase from Francisella tularensis facilitates catalysis.
J.Biol.Chem., 288, 2013
|
|
5U1H
 
 | | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa | | Descriptor: | (2R,6S)-2-amino-6-(carboxyamino)-7-{[(1R)-1-carboxyethyl]amino}-7-oxoheptanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa
To be published
|
|
4F78
 
 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Minasov, G, Egorova, O, Di Leo, R, Kudritska, M, Yim, V, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4FEX
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor tyrphostin AG1478 | | Descriptor: | ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, KANAMYCIN A, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4ECL
 
 | | Crystal structure of the cytoplasmic domain of vancomycin resistance serine racemase VanTg | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine racemase | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Cosme, J, Di Leo, R, Krishnamoorthy, M, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structural and Functional Adaptation of Vancomycin Resistance VanT Serine Racemases.
MBio, 6, 2015
|
|
