1US0
 
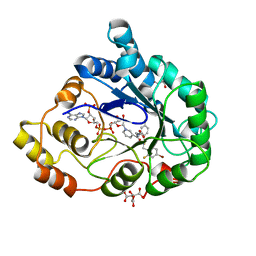 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | Authors: | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | Deposit date: | 2003-11-16 | | Release date: | 2004-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.66 Å) | | Cite: | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
8GEW
 
 | | H-FABP crystal soaked in a bromo palmitic acid solution | | Descriptor: | 2-Bromopalmitic acid, Fatty acid-binding protein, heart, ... | | Authors: | Howard, E, Cousido-Siah, A, Alvarez, A, Espinosa, Y, Podjarny, A, Mitschler, A, Carlevaro, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lipid exchange in crystal-confined fatty acid binding proteins: X-ray evidence and molecular dynamics explanation.
Proteins, 91, 2023
|
|
6B9P
 
 | | Structure of GH 38 Jack Bean alpha-mannosidase in complex with a 36-valent iminosugar cluster inhibitor | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{9-[4-(methoxymethyl)-1H-1,2,3-triazol-1-yl]nonyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase from Canavalia ensiformis (jack bean), ... | | Authors: | Howard, E, Cousido-Siah, A, Lepage, M, Bodlenner, A, Mitschler, A, Meli, A, De Riccardis, F, Izzo, I, Podjarny, A, Compain, P. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural Basis of Outstanding Multivalent Effects in Jack Bean alpha-Mannosidase Inhibition.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6B9O
 
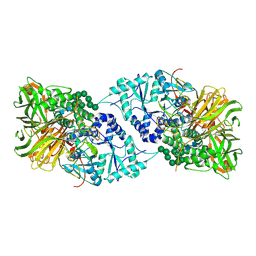 | | Structure of GH 38 Jack Bean alpha-mannosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase from Canavalia ensiformis (jack bean), ZINC ION, ... | | Authors: | Howard, E, Cousido-Siah, A, Lepage, M, Bodlenner, A, Mitschler, A, Meli, A, De Riccardis, F, Izzo, I, Podjarny, A, Compain, P. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structural Basis of Outstanding Multivalent Effects in Jack Bean alpha-Mannosidase Inhibition.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3QF6
 
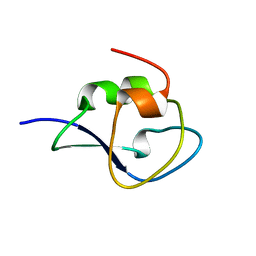 | | Neutron structure of type-III Antifreeze Protein allows the reconstruction of AFP-ice interface | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Howard, E.I, Blakeley, M.P, Haertlein, M, Petit-Haertlein, I, Mitschler, A, Fisher, S.J, Cousido-Siah, A, Salvay, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A. | | Deposit date: | 2011-01-21 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.85 Å) | | Cite: | Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface.
J.Mol.Recognit., 24, 2011
|
|
1DV0
 
 | |
1F4I
 
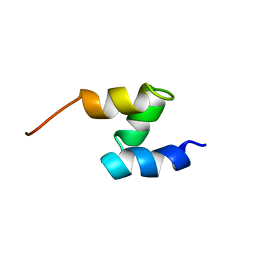 | | SOLUTION STRUCTURE OF THE HHR23A UBA(2) MUTANT P333E, DEFICIENT IN BINDING THE HIV-1 ACCESSORY PROTEIN VPR | | Descriptor: | UV EXCISION REPAIR PROTEIN PROTEIN RAD23 HOMOLOG A | | Authors: | Withers-Ward, E.S, Mueller, T.D, Chen, I.S, Feigon, J. | | Deposit date: | 2000-06-07 | | Release date: | 2000-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr.
Biochemistry, 39, 2000
|
|
1H5B
 
 | | T cell receptor Valpha11 (AV11S5) domain | | Descriptor: | CHLORIDE ION, GLYCEROL, MURINE T CELL RECEPTOR (TCR) VALPHA DOMAIN | | Authors: | Machius, M, Cianga, P, Deisenhofer, J, Sally Ward, E. | | Deposit date: | 2001-05-21 | | Release date: | 2001-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a T Cell Receptor Valpha11 (Av11S5) Domain: New Canonical Forms for the First and Second Complementarity Determining Regions
J.Mol.Biol., 310, 2001
|
|
1DJ2
 
 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
1K2D
 
 | | Crystal structure of the autoimmune MHC class II I-Au complexed with myelin basic protein 1-11 at 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 class II histocompatibility antigen, A-U alpha chain, ... | | Authors: | He, X.L, Radu, C, Ward, E.S, Garcia, K.C. | | Deposit date: | 2001-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshot of aberrant antigen presentation linked to autoimmunity: the
immunodominant epitope of MBP complexed with I-Au
IMMUNITY, 17, 2002
|
|
1DJ3
 
 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
1ZEG
 
 | | STRUCTURE OF B28 ASP INSULIN IN COMPLEX WITH PHENOL | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1ZEI
 
 | | CROSS-LINKED B28 ASP INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-07-14 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1ZEH
 
 | | STRUCTURE OF INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
5AMF
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment Ethyl 4,5,6,7- tetrahydro-1H-indazole-5-carboxylate (SGC - Diamond I04-1 fragment screening) | | Descriptor: | BROMODOMAIN-CONTAINING PROTEIN 1, ETHYL (5R)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE-5-CARBOXYLATE, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment Ethyl 4,5,6,7-Tetrahydro-1H-Indazole-5-Carboxylate
To be Published
|
|
5AME
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment 4-acetyl- piperazin-2-one (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 4-acetyl-piperazin-2-one, BROMODOMAIN-CONTAINING PROTEIN 1, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment 4-Acetyl-Piperazin-2-One
To be Published
|
|
4RVM
 
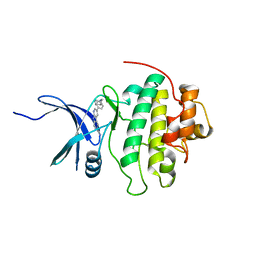 | | CHK1 kinase domain with diazacarbazole compound 19 | | Descriptor: | 3-[4-(piperidin-1-ylmethyl)phenyl]-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Gazzard, L, Blackwood, E, Burton, B, Chapman, K, Chen, H, Crackett, P, Drobnick, J, Ellwood, C, Epler, J, Flagella, M, Goodacre, S, Halladay, J, Hunt, H, Kintz, S, Lyssikatos, J, MacLeod, C, Ramiscal, S, Schmidt, S, Seward, E, Wiesmann, C, Williams, K, Wu, P, Yee, S, Yen, I, Malek, S. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
7Q46
 
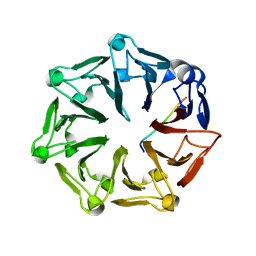 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Pericentriolar material 1 protein | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46002531 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein
To Be Published
|
|
7Q40
 
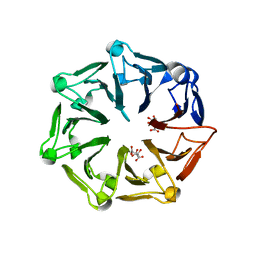 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35002232 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2
To Be Published
|
|
7Q44
 
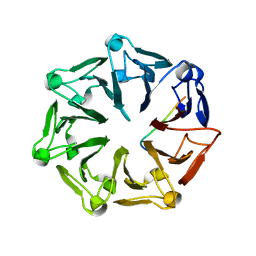 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of deubiquitinase USP35 | | Descriptor: | CITRIC ACID, Deubiquitinase USP35 peptide, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.20007777 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of deubiquitinase USP35
To Be Published
|
|
7Q41
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of ubiquitin-protein ligase E3A (E6AP) | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Ubiquitin-protein ligase E3A (E6AP) peptide | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.01478052 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of ubiquitin-protein ligase E3A (E6AP)
To Be Published
|
|
7Q45
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1 | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Myelin transcription factor 1 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09999585 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1
To Be Published
|
|
7Q42
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of chromatin reader BAZ2B | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, CITRIC ACID, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95002484 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of chromatin reader BAZ2B
To Be Published
|
|
7Q43
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10 | | Descriptor: | CITRIC ACID, Dedicator of cytokinesis protein 10 peptide, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40002346 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10
To Be Published
|
|
3UM9
 
 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Bpro0530 | | Descriptor: | Haloacid dehalogenase, type II, NICKEL (II) ION, ... | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-12 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
