3DB5
 
 | | Crystal structure of methyltransferase domain of human PR domain-containing protein 4 | | Descriptor: | PR domain zinc finger protein 4 | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of methyltransferase domain of human PR domain-containing protein 4.
To be Published
|
|
4GLM
 
 | | Crystal structure of the SH3 Domain of DNMBP protein [Homo sapiens] | | Descriptor: | Dynamin-binding protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Guan, X, Huang, H, Tempel, W, Gu, J, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SH3 Domain of DNMBP protein [Homo sapiens]
to be published
|
|
5IID
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 2-(3,4-dihydroxyphenyl)-5-hydroxy-4H-chromen-4-one | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5-hydroxy-4H-1-benzopyran-4-one, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S. | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
4PWY
 
 | | Crystal structure of a Calmodulin-lysine N-methyltransferase fragment | | Descriptor: | Calmodulin-lysine N-methyltransferase, GLYCEROL, MALONATE ION, ... | | Authors: | Tempel, W, Hong, B.S, Walker, J.R, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Calmodulin-lysine N-methyltransferase fragment
TO BE PUBLISHED
|
|
5II2
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, CITRIC ACID, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S. | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
2R6O
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Thiobacillus denitrificans | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative diguanylate cyclase/phosphodiesterase (GGDEF & EAL domains) | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-18 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
2R8W
 
 | | The crystal structure of dihydrodipicolinate synthase (Atu0899) from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETATE ION, AGR_C_1641p, CHLORIDE ION | | Authors: | Tan, K, Dong, A, Xu, X, Gu, J, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of dihydrodipicolinate synthase (Atu0899) from Agrobacterium tumefaciens str. C58.
To be Published
|
|
2RFI
 
 | | Crystal structure of catalytic domain of human euchromatic histone methyltransferase 1 in complex with SAH and dimethylated H3K9 peptide | | Descriptor: | Histone H3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
2RBC
 
 | | Crystal structure of a putative ribokinase from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-11-13 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a putative ribokinase from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
3LS8
 
 | | Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol | | Descriptor: | 3-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Kraulis, P, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, Van den Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol
To be Published
|
|
6DXH
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-tert-butylphenyl)-4-oxobutanoate | | Descriptor: | 4-(4-tert-butylphenyl)-4-oxobutanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Harding, R.J, Mann, M.K, Ravichandran, M, Ferreira de Freitas, R, Franzoni, I, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
3LWK
 
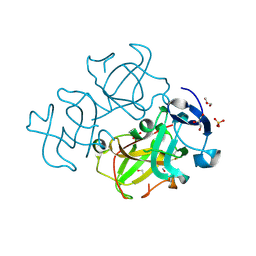 | | Crystal structure of human Beta-crystallin A4 (CRYBA4) | | Descriptor: | Beta-crystallin A4, GLYCEROL, PHOSPHATE ION | | Authors: | Chaikuad, A, Shafqat, N, Krojer, T, Yue, W.W, Cocking, R, Vollmar, M, Muniz, J.R.C, Pike, A.C.W, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Beta-crystallin A4 (CRYBA4)
To be Published
|
|
3LZK
 
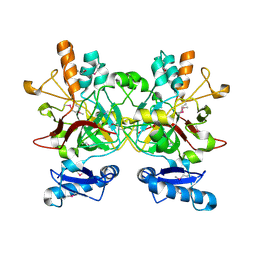 | | The crystal structure of a probably aromatic amino acid degradation proteiN from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, Fumarylacetoacetate hydrolase family protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a probably aromatic amino acid degradation protein from Sinorhizobium meliloti 1021
To be Published
|
|
3MA3
 
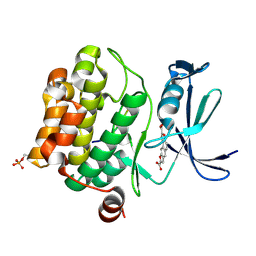 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a naphtho-difuran ligand | | Descriptor: | Pimtide, Proto-oncogene serine/threonine-protein kinase pim-1, naphtho[2,1-b:7,6-b']difuran-2,8-dicarboxylic acid | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Vollmar, M, von Delft, F, Cochet, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
3MEV
 
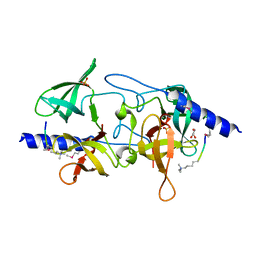 | | Crystal structure of SGF29 in complex with R2AK4me3 | | Descriptor: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | Authors: | Bian, C.B, Xu, C, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3MDY
 
 | | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Bone morphogenetic protein receptor type-1B, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chaikuad, A, Sanvitale, C, Mahajan, P, Daga, N, Cooper, C, Krojer, T, Alfano, I, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189
To be Published
|
|
3MEU
 
 | | Crystal structure of SGF29 in complex with H3R2me2sK4me3 | | Descriptor: | Histone H3, SAGA-associated factor 29 homolog, SULFATE ION | | Authors: | Bian, C.B, Xu, C, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3MHK
 
 | | Human tankyrase 2 - catalytic PARP domain in complex with 2-(2-pyridyl)-7,8-dihydro-5h-thiino[4,3-d]pyrimidin-4-ol | | Descriptor: | 2-pyridin-2-yl-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, GLYCEROL, Tankyrase-2, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Markova, N, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
4UYB
 
 | | Crystal structure of SEC14-like protein 3 | | Descriptor: | 1,2-ETHANEDIOL, SEC14-LIKE PROTEIN 3, UNKNOWN LIGAND | | Authors: | Kopec, J, Goubin, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Sec14-Like Protein 3
To be Published
|
|
4FU3
 
 | | CID of human RPRD1B | | Descriptor: | CHLORIDE ION, Regulation of nuclear pre-mRNA domain-containing protein 1B, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CID of human RPRD1B
TO BE PUBLISHED
|
|
5I3R
 
 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | Descriptor: | BMP-2-inducible protein kinase, N-[6-(3-{[(cyclopropylmethyl)sulfonyl]amino}phenyl)-1H-indazol-3-yl]cyclopropanecarboxamide, PHOSPHATE ION | | Authors: | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor
To Be Published
|
|
6EIM
 
 | | Human STK10 bound to GW683134A | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 10, ~{N}-[5-[4-[[2-fluoranyl-5-(trifluoromethyl)phenyl]carbamoylamino]phenoxy]-1~{H}-benzimidazol-2-yl]furan-2-carboxamide | | Authors: | Sorrell, F.J, Berger, B.-T, Salah, E, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Elkins, J.M. | | Deposit date: | 2017-09-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Human STK10 bound to GW683134
To Be Published
|
|
3MSE
 
 | | Crystal structure of C-terminal domain of PF110239. | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, putative, ... | | Authors: | Wernimont, A.K, Artz, J.D, Hutchinson, A, Sullivan, H, Weadge, J, Tempel, W, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Lin, Y.H, Neculai, A.M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of C-terminal domain of PF110239
To be Published
|
|
4Q0O
 
 | | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(3R)-3-phenylpiperidin-1-yl]prop-2-en-1-one, POTASSIUM ION, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand
To be Published
|
|
5IGM
 
 | | Crystal structure of the bromodomain of human BRD9 in complex with bromosporine (BSP) | | Descriptor: | Bromodomain-containing protein 9, Bromosporine | | Authors: | Tallant, C, Filippakopoulos, P, Picaud, S, Nunez-Alonso, G, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia.
Sci Adv, 2, 2016
|
|
