4QXA
 
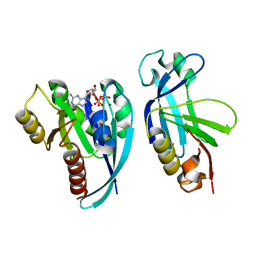 | | Crystal structure of the Rab9A-RUTBC2 RBD complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-9A, ... | | Authors: | Zhang, Z, Wang, S, Ding, J. | | Deposit date: | 2014-07-19 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Rab9A-RUTBC2 RBD complex reveals the molecular basis for the binding specificity of Rab9A with RUTBC2.
Structure, 22, 2014
|
|
7N3O
 
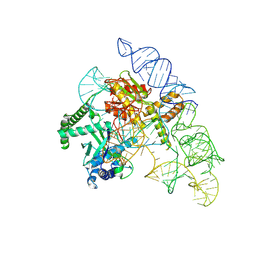 | | Cryo-EM structure of the Cas12k-sgRNA complex | | Descriptor: | Cas12k, Single guide RNA | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
7N3P
 
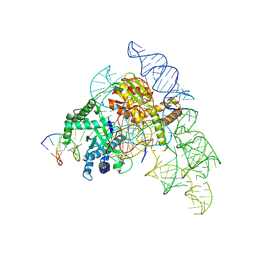 | | Cryo-EM structure of the Cas12k-sgRNA-dsDNA complex | | Descriptor: | Cas12k, DNA (5'-D(*CP*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*AP*AP*CP*CP*GP*AP*GP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*CP*TP*CP*GP*GP*TP*T)-3'), ... | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
8WX8
 
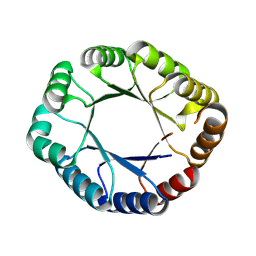 | |
1M07
 
 | | RESIDUES INVOLVED IN THE CATALYSIS AND BASE SPECIFICITY OF CYTOTOXIC RIBONUCLEASE FROM BULLFROG (RANA CATESBEIANA) | | Descriptor: | 5'-D(*AP*CP*GP*A)-3', Ribonuclease | | Authors: | Leu, Y.-J, Chern, S.-S, Wang, S.-C, Hsiao, Y.-Y, Amiraslanov, I, Liaw, Y.-C, Liao, Y.-D. | | Deposit date: | 2002-06-12 | | Release date: | 2003-01-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Residues involved in the catalysis, base specificity, and cytotoxicity of ribonuclease from Rana catesbeiana based upon mutagenesis and X-ray crystallography
J.Biol.Chem., 278, 2003
|
|
7T0X
 
 | | Structure of the larger diameter PSMalpha3 nanotube | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Kreutzberger, M.A, Wang, S, Beltran, L.C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-11-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SZZ
 
 | | Structure of the smaller diameter PSMalpha3 nanotubes | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Beltran, L.C, Kreutzberger, M.A, Wang, S, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-11-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T8U
 
 | |
2FEX
 
 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | Authors: | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
2RLI
 
 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
4TPT
 
 | | Crystal Structure of the Human LIMK2 Kinase Domain In Complex With a Non-ATP Competitive Inhibitor | | Descriptor: | LIM domain kinase 2, N-{4-[(1S)-1,2-dihydroxyethyl]benzyl}-N-methyl-4-(phenylsulfamoyl)benzamide | | Authors: | Goodwin, N.C, Cianchetta, G, Hamman, B.L, Burgoon, H.A, Healy, J, Mabon, S, Strobel, E.D, Wang, S, Rawlins, D.B. | | Deposit date: | 2014-06-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Type III Inhibitor of LIM Kinase 2 That Binds in a DFG-Out Conformation.
Acs Med.Chem.Lett., 6, 2015
|
|
6QUY
 
 | | NgCKK (N.Gruberi CKK) decorated 13pf taxol-GDP microtubule | | Descriptor: | CKK domain protein, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QVE
 
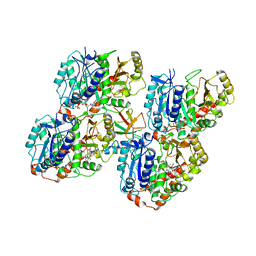 | | NgCKK (Naegleria Gruberi CKK) decorated 14pf taxol-GDP microtubule | | Descriptor: | Beta1-tubulin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
3H3M
 
 | | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica | | Descriptor: | Flagellar protein FliT, UNKNOWN | | Authors: | Shumilin, I.A, Wang, S, Chruszcz, M, Xu, X, Le, B, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-16 | | Release date: | 2009-04-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica
To be Published
|
|
7YH5
 
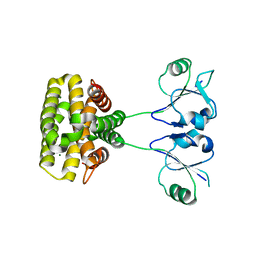 | | MazG(Mycobacterium tuberculosis) | | Descriptor: | MAGNESIUM ION, Nucleoside triphosphate pyrophosphohydrolase | | Authors: | Li, J, Wang, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis.
Front Microbiol, 14, 2023
|
|
8JX4
 
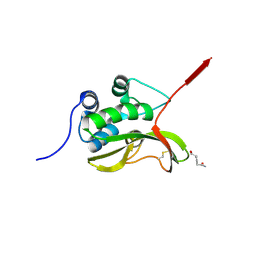 | | Structure of the catalytic domain of pseudomurein endo-isopeptidases PeiW | | Descriptor: | ALANINE, D-GLUTAMIC ACID, PeiW | | Authors: | Guo, L.Z, Zhao, N.L, Len, H, Wang, S.X, Cha, G.H, Bai, L.p, Bao, R. | | Deposit date: | 2023-06-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Structure and Function of Pseudomurein Endoisopeptidases: peiW and peiP
To Be Published
|
|
4ZN9
 
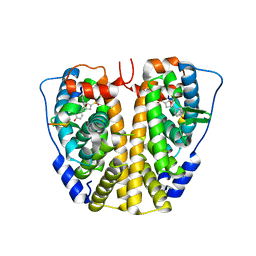 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Oxabicyclic Heptene Sulfonate (OBHS) | | Descriptor: | Estrogen receptor, Nuclear receptor-interacting peptide, cyclohexa-2,5-dien-1-yl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands.
Chemmedchem, 7, 2012
|
|
8W6K
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BL03HB: a Laue microdiffraction beamline for both protein crystallography and materials science at SSRF
Nucl.Sci.Tech., 35, 2024
|
|
8FD2
 
 | | Cryo-EM structure of Cascade complex in type I-B CAST system | | Descriptor: | A Type I-B CRISPR-associated protein Cas5, RNA, Type I-B CRISPR-associated protein Cas11, ... | | Authors: | Chang, L, Wang, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector.
Cell, 186, 2023
|
|
8FCJ
 
 | |
8FD3
 
 | |
8FF5
 
 | |
7YS6
 
 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | Descriptor: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | Deposit date: | 2022-08-11 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1SFK
 
 | | Core (C) protein from West Nile Virus, subtype Kunjin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Core protein, ... | | Authors: | Dokland, T, Walsh, M, Mackenzie, J.M, Khromykh, A.A, Ee, K.-H, Wang, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | West nile virus core protein; tetramer structure and ribbon formation
Structure, 12, 2004
|
|
3DCA
 
 | | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target | | Descriptor: | RPA0582, SULFATE ION | | Authors: | Sledz, P, Wang, S, Chruszcz, M, Yim, V, Kudritska, M, Evdokimova, E, Turk, D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target
To be Published
|
|
