8XVT
 
 | | The core subcomplex of human NuA4/TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVG
 
 | | Structure of human NuA4/TIP60 complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
5BR3
 
 | | Crystal structure of hemagglutinin of A/Taiwan/2/2013 (H6N1) in complex with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Studies of Influenza Virus A/H6 Hemagglutinin.
Plos One, 7, 2015
|
|
4M40
 
 | | Crystal structure of hemagglutinin of influenza virus B/Yamanashi/166/1998 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structural basis for the divergent evolution of influenza B virus hemagglutinin.
Virology, 446, 2013
|
|
5BQZ
 
 | | Crystal structure of hemagglutinin of A/Chicken/Guangdong/S1311/2010 (H6N6) in complex with human-like receptor LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 CHAIN, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2015-05-29 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural and Functional Studies of Influenza Virus A/H6 Hemagglutinin.
Plos One, 10, 2015
|
|
4M44
 
 | | Crystal structure of hemagglutinin of influenza virus B/Yamanashi/166/1998 in complex with avian-like receptor LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the divergent evolution of influenza B virus hemagglutinin.
Virology, 446, 2013
|
|
5BR6
 
 | | Crystal structure of hemagglutinin of A/Taiwan/2/2013 (H6N1) in complex with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Studies of Influenza Virus A/H6 Hemagglutinin.
Plos One, 7, 2015
|
|
5BNY
 
 | | Crystal structure of hemagglutinin of A/Chicken/Guangdong/S1311/2010 (H6N6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural and Functional Studies of Influenza Virus A/H6 Hemagglutinin.
Plos One, 7, 2015
|
|
5BR0
 
 | | Crystal structure of hemagglutinin of A/Taiwan/2/2013 (H6N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Functional Studies of Influenza Virus A/H6 Hemagglutinin.
Plos One, 7, 2015
|
|
5BQY
 
 | | Crystal structure of hemagglutinin of A/Chicken/Guangdong/S1311/2010 (H6N6) in complex with avian-like receptor LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and Functional Studies of Influenza Virus A/H6 Hemagglutinin.
Plos One, 7, 2015
|
|
8ZLE
 
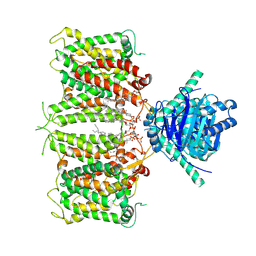 | | hAE3NTD2TMD with PT5,CLR, and Y01 | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate, ... | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-05-19 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8Y85
 
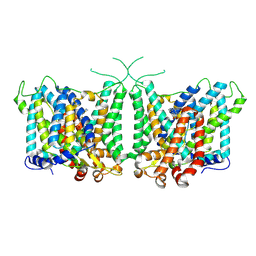 | | Human AE3 with NaHCO3- and DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
7C01
 
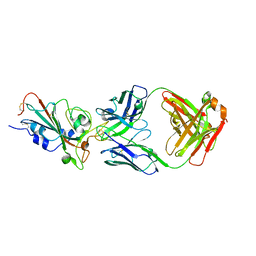 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
1KAK
 
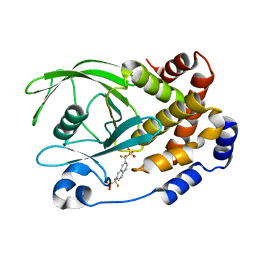 | | Human Tyrosine Phosphatase 1B Complexed with an Inhibitor | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1, {[7-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALEN-2-YL]-DIFLUORO-METHYL}-PHOSPHONIC ACID | | Authors: | Jia, Z, Ye, Q, Dinaut, A.N, Wang, Q, Waddleton, D, Payette, P, Ramachandran, C, Kennedy, B, Hum, G, Taylor, S.D. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of protein tyrosine phosphatase 1B in complex with inhibitors bearing two phosphotyrosine mimetics.
J.Med.Chem., 44, 2001
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
6VJT
 
 | |
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
