7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
 | |
7F9Z
 
 | | GHRP-6-bound ghrelin receptor in complex with Gq | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Zhuang, Y, Xu, P, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-07-05 | | Release date: | 2021-08-18 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor.
Nat Commun, 12, 2021
|
|
4ESP
 
 | | Crystal Structure of Peanut Allergen Ara h 5 | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Profilin | | Authors: | Wang, Y, Zhang, Y.Z. | | Deposit date: | 2012-04-23 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Peanut ( Arachis hypogaea ) Allergen Ara h 5.
J.Agric.Food Chem., 61, 2013
|
|
1R6K
 
 | | HPV11 E2 TAD crystal structure | | Descriptor: | HPV11 REGULATORY PROTEIN E2 | | Authors: | Wang, Y, Coulombe, R. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the E2 Transactivation Domain of Human Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
3FBD
 
 | |
5ULT
 
 | | HIV-1 wild Type protease with GRL-100-13A (a Crown-like Oxotricyclic Core as the P2-Ligand with the sulfonamide isostere as the P2' group) | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and Development of Highly Potent HIV-1 Protease Inhibitors with a Crown-Like Oxotricyclic Core as the P2-Ligand To Combat Multidrug-Resistant HIV Variants.
J. Med. Chem., 60, 2017
|
|
3K61
 
 | | Crystal structure of FBF-2/fog-1 FBEa complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*AP*AP*AP*UP*C)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
179D
 
 | |
186D
 
 | |
4IW2
 
 | | HSA-glucose complex | | Descriptor: | D-glucose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Luo, Z, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
6JOH
 
 | |
4IW1
 
 | | HSA-fructose complex | | Descriptor: | D-fructose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
8JKE
 
 | | AfsR(T337A) transcription activation complex | | Descriptor: | DNA(65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wang, Y, Zheng, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural and functional characterization of AfsR, an SARP family transcriptional activator of antibiotic biosynthesis in Streptomyces.
Plos Biol., 22, 2024
|
|
4P6G
 
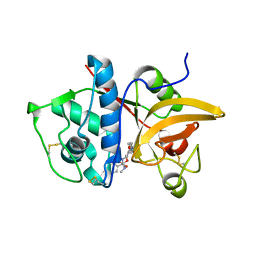 | | Crystal Structure of Human Cathepsin S Bound to a Non-covalent Inhibitor. | | Descriptor: | (3R,4S)-4-[(4-fluorobenzoyl)amino]-6-[4-(oxetan-3-yl)piperazin-1-yl]-3,4-dihydro-2H-chromen-3-yl methylcarbamate, Cathepsin S | | Authors: | Wang, Y, Jadhav, P.K. | | Deposit date: | 2014-03-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of Cathepsin S Inhibitor LY3000328 for the Treatment of Abdominal Aortic Aneurysm.
Acs Med.Chem.Lett., 5, 2014
|
|
4JFK
 
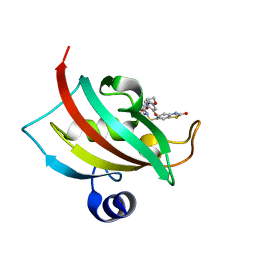 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
7CB7
 
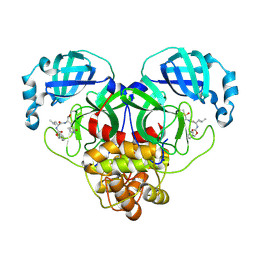 | | 1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y, Hung, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
3LJQ
 
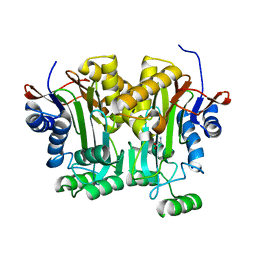 | |
5W87
 
 | |
5XKF
 
 | | Crystal structure of T2R-TTL-MPC6827 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
5XKE
 
 | | Crystal structure of T2R-TTL-Demecolcine complex | | Descriptor: | (7S)-1,2,3,10-tetramethoxy-7-(methylamino)-6,7-dihydro-5H-benzo[a]heptalen-9-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
4P6E
 
 | | Crystal Structure of Human Cathepsin S Bound to a Non-covalent Inhibitor | | Descriptor: | Cathepsin S, N-[(8R)-8-(benzoylamino)-5,6,7,8-tetrahydronaphthalen-2-yl]-4-methylpiperazine-1-carboxamide, SULFATE ION | | Authors: | Wang, Y, Jadhav, P.K, Deng, G.G. | | Deposit date: | 2014-03-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cathepsin S Inhibitor LY3000328 for the Treatment of Abdominal Aortic Aneurysm.
Acs Med.Chem.Lett., 5, 2014
|
|
1CW6
 
 | | REFINED SOLUTION STRUCTURE OF LEUCOCIN A | | Descriptor: | TYPE IIA BACTERIOCIN LEUCOCIN A | | Authors: | Wang, Y, Henz, M.E, Gallagher, N.L.F, Chai, S, Yan, L.Z, Gibbs, A.C, Stiles, M.E, Wishart, D.S, Vederas, J.C. | | Deposit date: | 1999-08-25 | | Release date: | 1999-09-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria.
Biochemistry, 38, 1999
|
|
3K62
 
 | | Crystal structure of FBF-2/gld-1 FBEb complex | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*UP*AP*UP*C)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4PES
 
 | |
