6XL8
 
 | | Crystal structure of 3-O-Sulfotransferase isoform 3 in complex with 8mer oligosaccharide with no 6S sulfation | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 3A1, IODIDE ION, ... | | Authors: | Pedersen, L.C, Liu, J, Wander, R. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Deciphering the substrate recognition mechanisms of the heparan sulfate 3- O -sulfotransferase-3.
Rsc Chem Biol, 2, 2021
|
|
5FB8
 
 | | Structure of Interleukin-16 bound to the 14.1 antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-IL-16 antibody 14.1 Fab domain Heavy Chain, ... | | Authors: | Hall, G, Cowan, R, Bayliss, R, Carr, M. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a Potential Therapeutic Antibody Bound to Interleukin-16 (IL-16): MECHANISTIC INSIGHTS AND NEW THERAPEUTIC OPPORTUNITIES.
J.Biol.Chem., 291, 2016
|
|
5TRG
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide DPLG-2 | | Descriptor: | N,N-diethyl-N~2~-[(2E)-3-phenylprop-2-enoyl]-L-asparaginyl-4-fluoro-N-[(naphthalen-1-yl)methyl]-L-phenylalaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, R.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TS0
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2208 | | Descriptor: | (2S)-N-{(2S)-3-methoxy-1-[(naphthalen-1-ylmethyl)amino]-1-oxopropan-2-yl}-4-oxo-2-[(3-phenylpropanoyl)amino]-4-(1H-pyrrol-1-yl)butanamide (non-preferred name), Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84679747 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRS
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2144 | | Descriptor: | N-tert-butoxy-N~2~-(5-methyl-1,2-oxazole-3-carbonyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.083567 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
4RIA
 
 | | FAN1 Nuclease bound to 5' phosphorylated nicked DNA | | Descriptor: | BARIUM ION, DNA (5'-D(*TP*TP*TP*TP*TP*TP*G*AP*GP*GP*CP*GP*TP*G)-3'), DNA (5'-D(P*AP*GP*AP*CP*TP*CP*CP*TP*C)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
4RID
 
 | | Human FAN1 nuclease | | Descriptor: | Fanconi-associated nuclease 1 | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
4RIC
 
 | | FAN1 Nuclease bound to 5' hydroxyl (dT-dT) single flap DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*GP*GP*AP*GP*TP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*CP*CP*AP*CP*GP*CP*CP*TP*AP*GP*AP*CP*TP*CP*CP*TP*C)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
4RI9
 
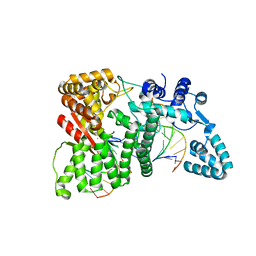 | |
4RIB
 
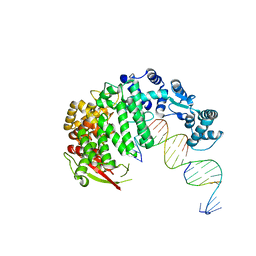 | | FAN1 Nuclease bound to 5' phosphorylated p(dT) single flap DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*GP*GP*AP*GP*TP*CP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*TP*GP*AP*GP*GP*CP*GP*TP*G)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
4RI8
 
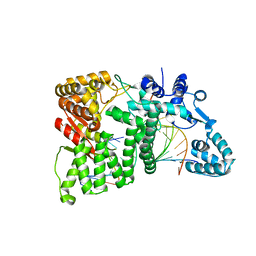 | |
5TRR
 
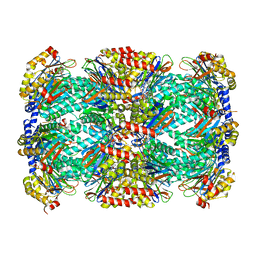 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2169 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-N-[(naphthalen-1-yl)methyl]-L-alaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRY
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2206 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-3-methoxy-1-(naphthalen-1-ylmethylamino)-1-oxidanylidene-propan-2-yl]-4-oxidanylidene-2-(3-phenylpropanoylamino)-4-piperidin-1-yl-butanamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.000008 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
1VJY
 
 | | Crystal Structure of a Naphthyridine Inhibitor of Human TGF-beta Type I Receptor | | Descriptor: | 2-[5-(6-METHYLPYRIDIN-2-YL)-2,3-DIHYDRO-1H-PYRAZOL-4-YL]-1,5-NAPHTHYRIDINE, TGF-beta receptor type I | | Authors: | Gellibert, F, Woolven, J, Fouchet, M.-H, Mathews, N, Goodland, H, Lovegrove, V, Laroze, A, Nguyen, V.-L, Sautet, S, Wang, R, Janson, C, Smith, W, Krysa, G, Boullay, V, de Gouville, A.-C, Huet, S, Hartley, D. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 1,5-Naphthyridine Derivatives as a Novel Series of Potent and Selective TGF-beta Type I Receptor Inhibitors.
J.Med.Chem., 47, 2004
|
|
7ESF
 
 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
7OXN
 
 | | Crystal Structure of TAP01 in complex with cyclised amyloid beta peptide | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta precursor protein, TAP01 family antibody heavy chain, ... | | Authors: | Hall, G, Cowan, R, Carr, M.D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a novel pseudo beta-hairpin structure of N-truncated amyloid-beta for use as a vaccine against Alzheimer's disease.
Mol Psychiatry, 27, 2022
|
|
7OW1
 
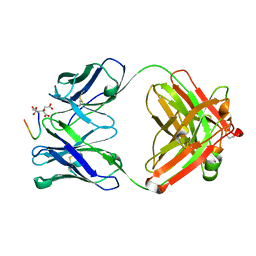 | | Crystal Structure of TAP01 in complex with amyloid beta peptide | | Descriptor: | Amyloid-beta precursor protein, CITRATE ANION, TAP01 family antibody heavy chain, ... | | Authors: | Hall, G, Cowan, R, Carr, M.D. | | Deposit date: | 2021-06-16 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a novel pseudo beta-hairpin structure of N-truncated amyloid-beta for use as a vaccine against Alzheimer's disease.
Mol Psychiatry, 27, 2022
|
|
6OY1
 
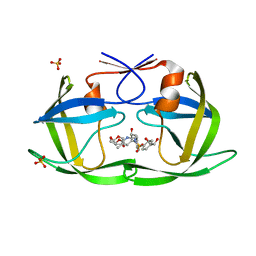 | | HIV-1 Protease NL4-3 WT in Complex with LR2-26 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-ethylbutyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXO
 
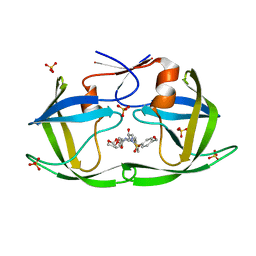 | | HIV-1 Protease NL4-3 WT in Complex with LR2-91 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXP
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass3 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXS
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-76 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[({4-[(1R)-1-hydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
2KZX
 
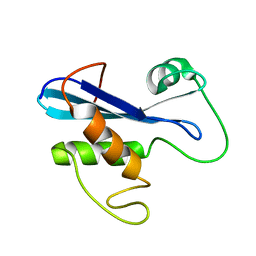 | | Solution NMR Structure of A3DHT5 from Clostridium thermocellum, Northeast Structural Genomics Consortium Target CmR116 | | Descriptor: | Uncharacterized protein | | Authors: | Mills, J.L, Eletsky, A, Lee, H, Janjua, H, Ciccosanti, C, Wang, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target CmR116
To be Published
|
|
3I4H
 
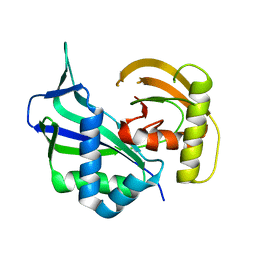 | | Crystal structure of Cas6 in Pyrococcus furiosus | | Descriptor: | endoribonuclease | | Authors: | Carte, J, Wang, R, Li, H, Terns, R.M, Terns, M.P. | | Deposit date: | 2009-07-01 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes
Genes Dev., 22, 2008
|
|
7C6Q
 
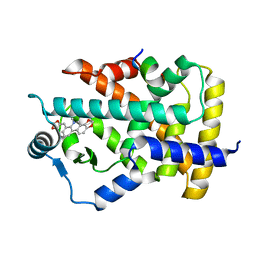 | | Novel natural PPARalpha agonist with a unique binding mode | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Peroxisome proliferator-activated receptor alpha | | Authors: | Tian, S.Y, Wang, R, Zheng, W.L, Li, Y. | | Deposit date: | 2020-05-22 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Basis for PPARs Activation by The Dual PPAR alpha / gamma Agonist Sanguinarine: A Unique Mode of Ligand Recognition.
Molecules, 26, 2021
|
|
1BBY
 
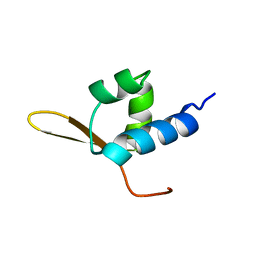 | | DNA-BINDING DOMAIN FROM HUMAN RAP30, NMR, MINIMIZED AVERAGE | | Descriptor: | RAP30 | | Authors: | Groft, C.M, Uljon, S.N, Wang, R, Werner, M.H. | | Deposit date: | 1998-04-26 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
